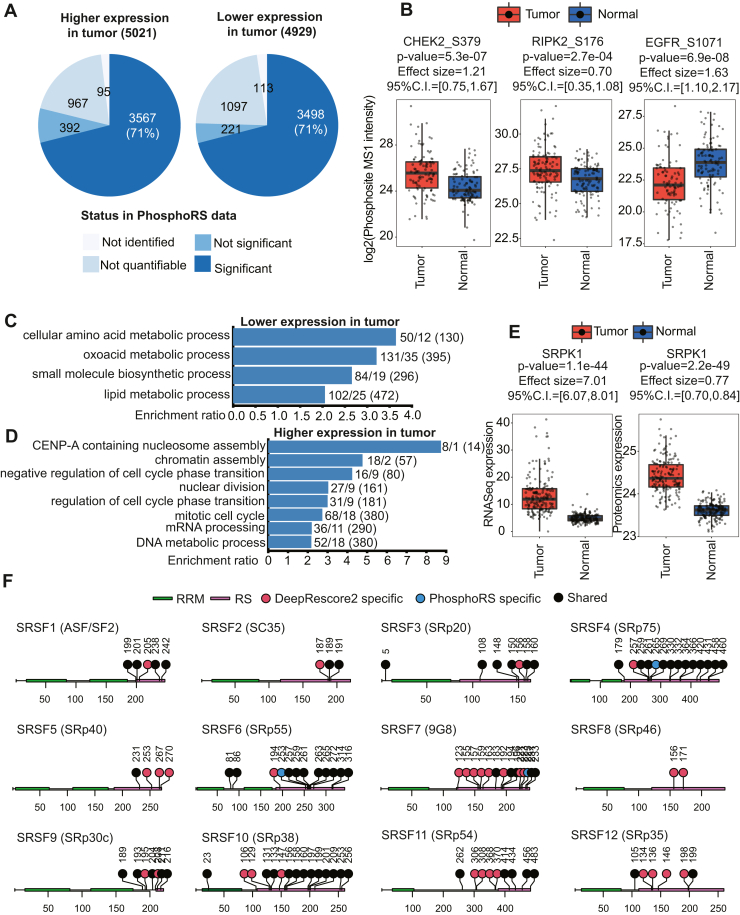
Fig. 6.
Differentially expressed phosphosites between tumor and NAT samples in the liver cancer dataset.A, the numbers of significantly altered (FDR < 0.01) phosphosites identified in DeepRescore2-derived data and their status in the analysis of PhosphoRS-derived data. B, comparison of phosphosite abundance in tumors and NATs for CHEK2_S379, RIPK2_S176, and EGFR_S1071, respectively. C, gene ontology enrichment analysis results of the parent proteins of the 4929 significantly down-regulated phosphosites. D, gene ontology enrichment analysis results of the parent proteins of the 5021 significantly up-regulated phosphosites. In C and D, the numbers at the end of each bar represent the number of parent proteins identified by both PhosphoRS and DeepRescore2, the number of parent proteins identified by DeepRescore2 alone, and the total number of parent proteins in the pathway, respectively. E, comparison of SRPK1 abundance between tumors and NATs at mRNA and protein levels, respectively. F, differentially expressed phosphosites detected in DeepRescore2-or PhosphoRS-derived data on 12 SR proteins. p values are based on the Wilcoxon rank sum test.