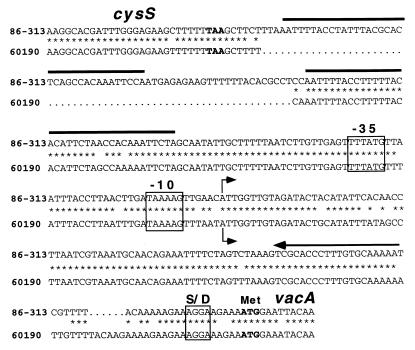
FIG. 2.
Comparison of cysS-vacA intergenic regions in H. pylori 86-313 (Tox−) and 60190 (Tox+). The cysS-vacA intergenic region from H. pylori 86-313 was PCR amplified and sequenced as described previously (14), and the sequence of the corresponding region from H. pylori 60190 has been reported previously (10). Analysis of the aligned sequences demonstrated a 63-bp insertion in the cysS-vacA intergenic region of Tox− strain 86-313. The corresponding absence of this sequence in Tox+ H. pylori 60190 is denoted by dots. Positions of nucleotide identity are denoted by asterisks. A 36-bp sequence and its direct repeat are indicated by solid bars. The vacA transcriptional start points (determined by primer extension analysis [Fig. 1]) are indicated by the bent arrows. The putative Shine-Dalgarno (S/D) sequence and putative −10 and −35 hexamers are boxed. The stop codon (TAA) of cysS and the start codon (ATG) of vacA are in boldface. The primer used to determine vacA transcriptional start points (Fig. 1) is indicated by an arrow over the complementary sequences.