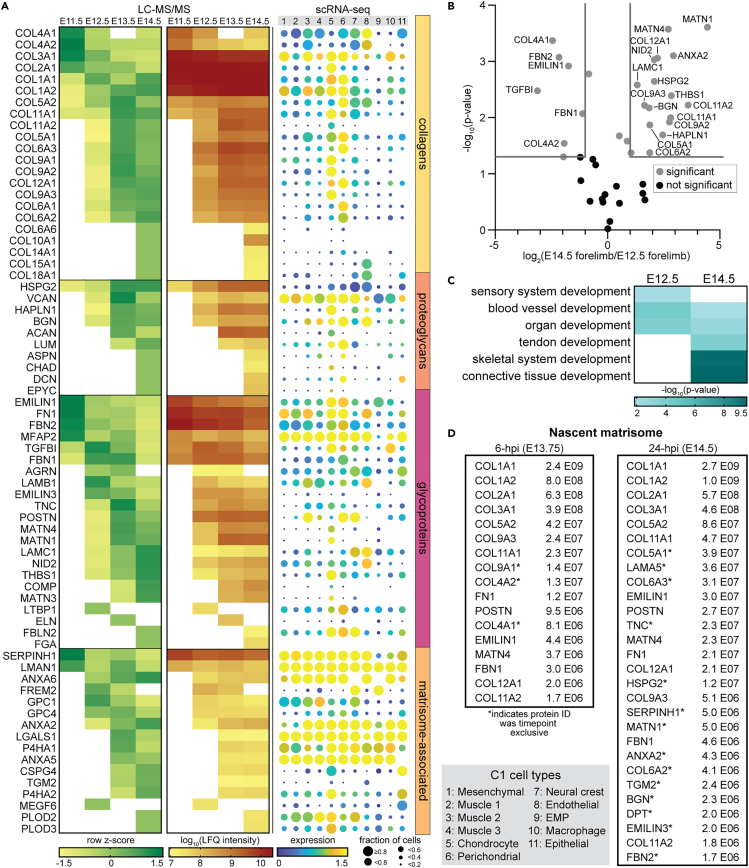
Figure 2.
ECM protein composition varies as a function of murine musculoskeletal development
(A, left) LFQ intensities of the matrisome composition of E11.5-E14.5 WT forelimbs were normalized and combined from both CS and IN fractions for each timepoint (Table S2). Row z-scores were calculated and averaged across biological replicates (n = 3) and proteins were clustered first by matrisome classification, then by relative abundance starting at E11.5. (A, center) Corresponding log10-transformed LFQ intensities for proteins shown in (A, left). White boxes signify proteins not identified in n ≥ 2 biological replicates. (A, right) C1 Fluidigm single-cell RNA-sequencing (scRNA-seq) data of the matrisome identified in E10-E15.5 limbs (Table S3), generated by He et al.3 Color indicates the log2(average expression +1) and circle size represents the percentage of cells expressing that transcript. Cell types defined by He et al.3 See Figure S2 for similar dot plot generated with 10X scRNA-seq data.3
(B) Many ECM proteins were significantly different in abundance between E12.5 and E14.5 forelimbs. Gray lines denote >2-fold change and p < 0.05 (two-tailed t-test).
(C) GO analysis of ECM proteins more abundant in, or exclusive to, E12.5 or E14.5 forelimbs generated distinct developmental terms.
(D) In vivo Aha-tagging (BONCAT), tissue fractionation, and enrichment of Aha-labeled ECM proteins were combined with LC-MS/MS analysis to identify the nascent matrisome in embryonic forelimbs (Table S2). Newly synthesized proteins (NSPs) that were part of the matrisome, identified in embryonic forelimbs 6- or 24-h post injection (hpi) of Aha at E13.5, were ranked in order of abundance. Proteins exclusively identified at E13.75 (6-hpi) or E14.5 (24-hpi) were denoted (∗) and the values displayed report the average raw protein intensity (n = 3).