Figure 6.
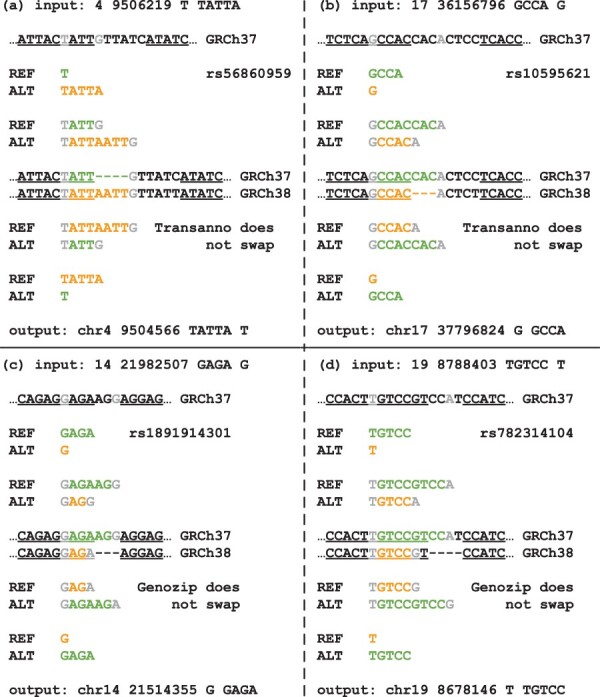
Examples of different liftover scenarios with discordance between BCFtools/liftover and either Transanno/liftvcf (a, b) or Genozip/DVCF (c, d) where the latter tools are unable to recognize the need for a swap of the reference with the alternate allele in the indel record. In each scenario, the 3’ anchor fails to map to the new assembly due to a small gap in the chain outlining the alignment between the two assemblies. BCFtools/liftover uses the Needleman-Wunsh algorithm to realign the sequence overlapping the gap and assess which base pair in the new assembly should act as a 3’ anchor. Underlined base pairs are base pairs covered by the hg19ToHg38.over.chain.gz chain file. Gray base pairs are 5’ and 3’ anchors for the maximally extended representations of the records.
