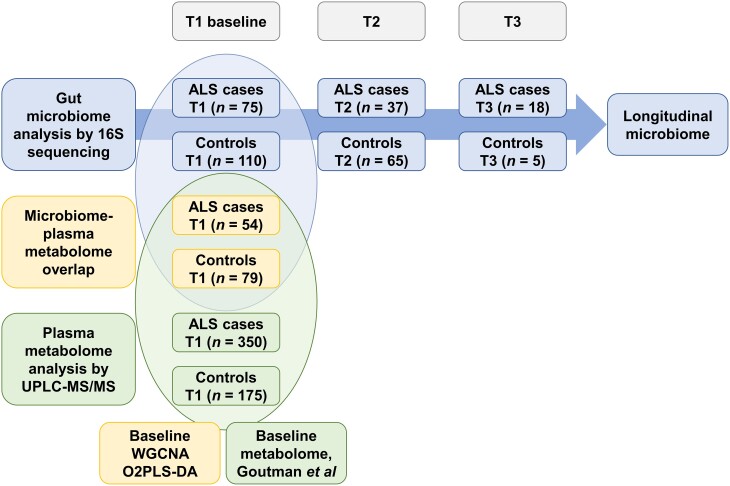
Figure 1.
Study design. Top: Gut microbiome was longitudinally profiled in faecal samples from amyotrophic lateral sclerosis (ALS) (n = 75) versus control (n = 110) participants by 16S rRNA sequencing at baseline (T1), T2 and T3. Bottom: Plasma metabolites were profiled in plasma samples from ALS (n = 350) versus control (n = 175) participants by untargeted metabolomics using ultra-high-performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) at baseline (T1), published in Goutman et al.15,16Middle: Fifty-three ALS cases and 80 controls overlap between the baseline microbiome and metabolome cohorts. Significantly associated bacteria and circulating metabolites were identified by weighted gene co-expression network analysis (WGCNA) and two-way orthogonal partial least square with discriminant analysis (O2PLS-DA).