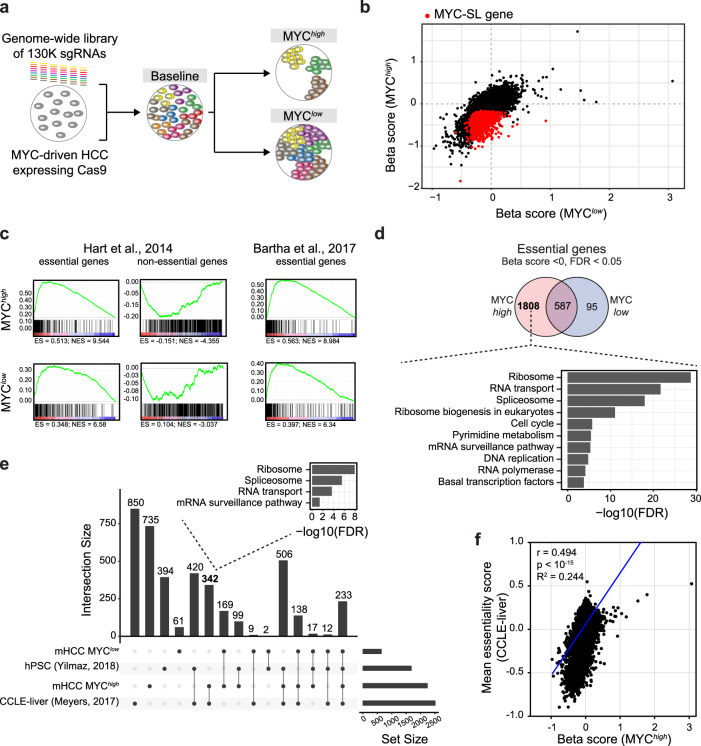
Fig. 1. CRISPR-based genome-wide screen identifies essential genes in MYC-driven HCC.
a Design of the CRISPR-based screen to identify vulnerabilities in MYC-driven HCC. A genome-wide sgRNA library (colored bars) is introduced in Cas9-expressing MYC-driven HCC cells to generate a baseline cell pool. Cells are then cultured under MYChigh or MYClow conditions resulting in a MYC synthetic lethal (MYC-SL) cell pool and a control cell pool, respectively. Deep sequencing of sgRNAs identifies genes that are essential only when MYC is highly expressed but not essential when MYC transgene expression is shut off. b Scatter plot of Beta scores (gene essentiality scores) in the control (MYClow; x-axis) versus MYC-SL (MYChigh; y-axis) condition. Loss of genes highlighted in red significantly reduced fitness of MYChigh HCC cells but had no significant effect on MYClow control cells. c Gene set enrichment analysis (GSEA) of essential and non-essential genes. Left and middle: Hart et al. Mol Syst Biol, 2014; Right: Bartha et al. Nat Rev Genet, 2017. d Venn diagram of essential genes identified in EC4 cells with high and low MYC levels. KEGG pathway analysis of MYC-SL genes (only essential in MYChigh condition) is shown. e Comparison of essential gene hits between our genome-wide CRISPR screen (MYChigh and MYClow control conditions) and essential genes identified in 13 human HCC cell lines (DepMap, CCLE-liver) and human pluripotent stem cells (hPSC). UpSet plot and numbers of overlapping essential genes between indicated genome-wide CRISPR screens are shown. f Correlation analysis of gene essentiality scores from our genome-wide CRISPR screen in MYC-driven murine HCC and essentiality scores in human HCC cell lines (DepMap, CCLE-liver). Pearson’s correlation coefficient (r), two-tailed p-value, and R2 of linear regression (blue line) are shown. For this figure, source data are provided as a Source Data file.