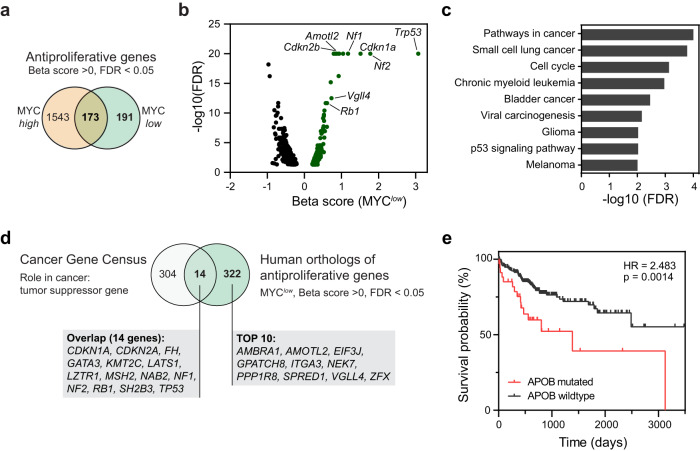
Fig. 2. Genome-wide CRISPR screen identifies tumor suppressor genes.
a Venn diagram of genes with antiproliferative function in MYChigh and MYClow conditions. b Volcano plot of Beta score versus significance (-log10(FDR)) of genes with FDR < 0.05 in the MYClow condition. Genes with antiproliferative properties (significant positive Beta score) are highlighted in green. Exemplary tumor suppressor genes are indicated. c KEGG pathway analysis of top 50 genes with significant positive Beta score in the MYClow condition. d Comparison of known tumor suppressor genes (TSGs) and human orthologues of genes with antiproliferative function in the MYClow condition. Known TSGs were derived from the Cancer Gene Census (https://cancer.sanger.ac.uk/cosmic/census). e Survival analysis of TCGA-LIHC assessing the effect of APOB mutation status on survival probability in HCC (p-value and hazard ratio (HR) from logrank test). For this figure, source data are provided as a Source Data file.