Figure 3.
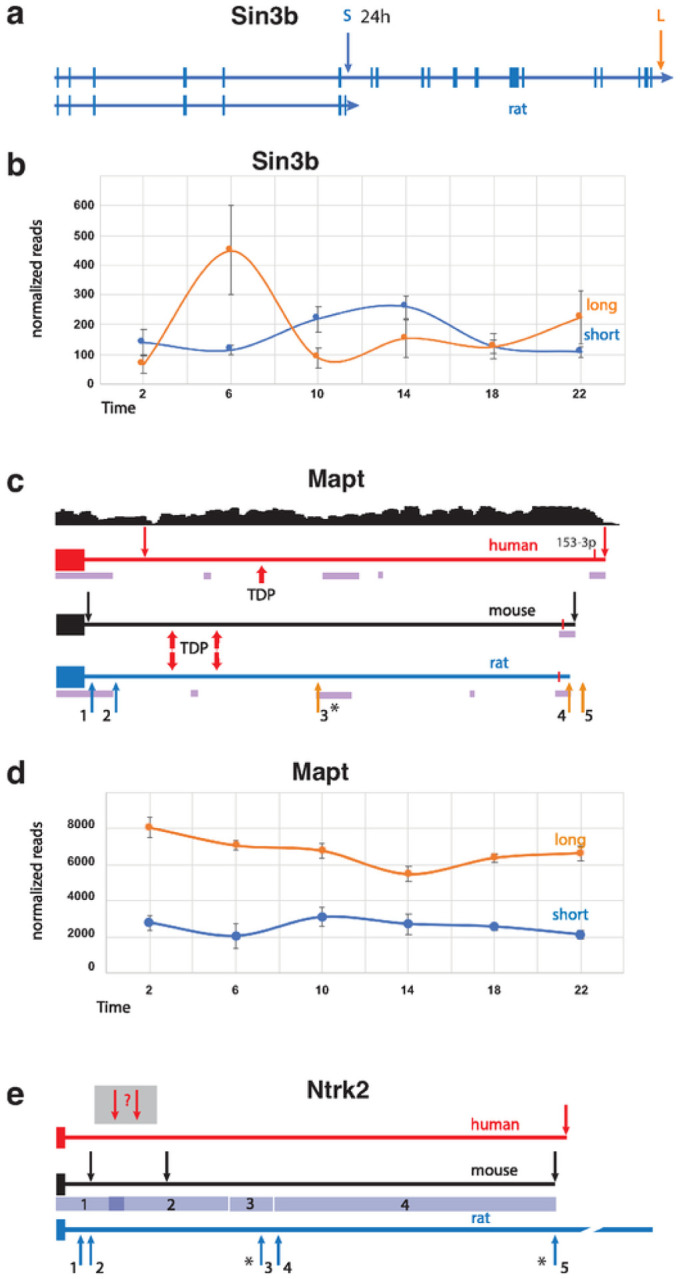
The Sin3b gene and 3’ UTR regions of the Mapt and Ntrk2 genes. (a) A map of the entire rat Sin3b gene depicts exons, introns and short and long APA sites. The corresponding genes in mouse and human are extremely similar. (b) The average normalized read counts ±SE (y-axis) of the short (circadian) and long Sin3b APAs are plotted against time-of-day (x-axis). (c) Maps of the 3’ UTR regions of the human, mouse, and rat Mapt genes are shown. Dark blue arrows indicate the positions of APA sites. In human MAPT, APA usage correlates with several brain disorders. RNA-seq coverage from individuals homozygous for the less common SNP allele that is associated with longer transcripts (adapted from Cui. et al.27) is shown above the human MAPT 3’ UTR map. Binding sites for TDP-43 (indicated by red arrows) that were experimentally determined in mouse align with putative sites in the rat gene, and one possible TDP-43 binding site is indicated in the human 3’UTR. The significantly circadian APA is marked with an asterisk. Blocks of homologous sequence between the rat and human genes that were found by BLAST search are indicated by purple bars. The 3’ UTR lengths are 4,380, 4,119 and 3,946 n.t. for human, mouse, and rat, respectively. (d) The average normalized read counts ±SE (y-axis) of the short Mapt isoforms lacking TDP binding sites (1+2) and the sum of the three longer isoforms (3+4+5) plotted against time-of-day (x-axis) are shown. (e) The 3’ UTR of tyrosine kinase-deficient (TK-) isoforms of the human, mouse, and rat Ntrk2 TK- genes are shown. Arrows indicate the positions of APA sites. The depicted rat APAs are from this current dataset. Circadian rat APAs are indicated with asterisks. The 3’ UTR lengths are 5,125, 5,008 and 8,004 n.t. for human, mouse, and rat, respectively. Mouse and rat sequence comparison by BLAST produced 4 segments having 91%, 83%, 86% and 82% identity for regions 1, 2, 3 and 4, depicted by blue bars.
