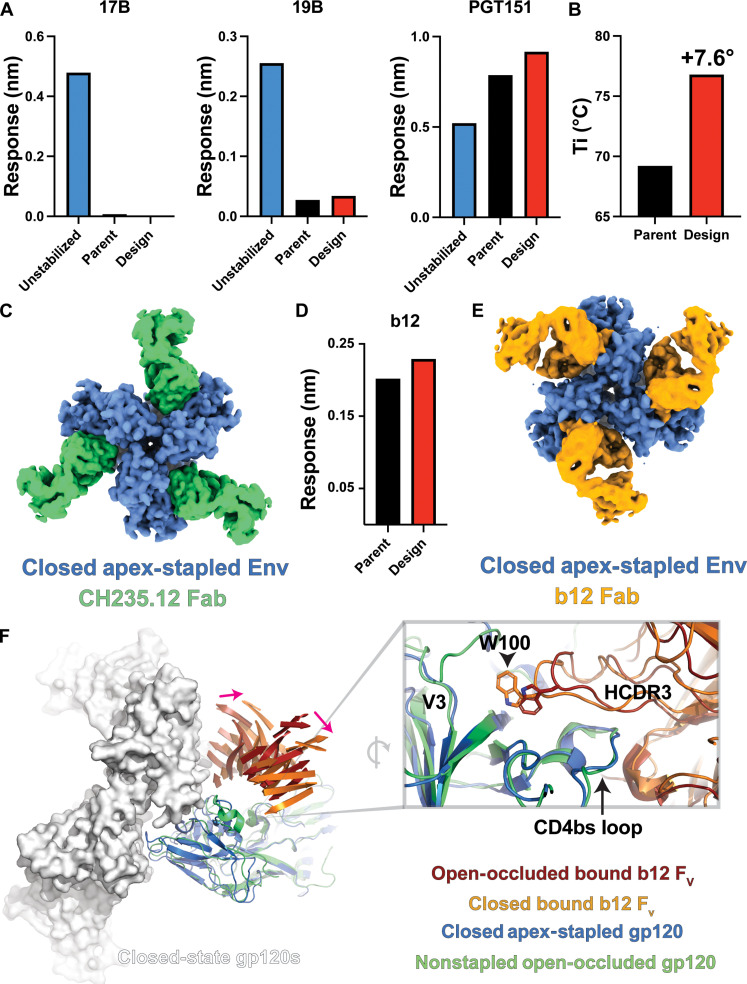
Fig. 5. Interprotomer disulfide bonds stabilize the closed Env trimer.
(A) Binding responses for a CH505 SOSIP, CH505 parent SOSIP containing previously identified stabilizing mutations, and the interprotomer disulfide-stapled CH505 SOSIP design interacting with the co-receptor binding 17b, V3 loop binding 19b, and the closed-state apex-interactive, trimer-specific PGT145 mAbs. (B) Differential fluorescence thermal denaturation inflection point temperature for the parent CH505 SOSIP and the interprotomer disulfide-stapled CH505 SOSIP. (C) Gaussian filtered map of the interprotomer disulfide-stapled CH505 SOSIP design bound to the CH235.12 Fab. (D) Binding responses for PGT151 captured stabilized CH505 parent SOSIP and interprotomer disulfide-stapled CH505 SOSIP design interacting with the b12 Fab. (E) Gaussian filtered map of the interprotomer disulfide-stapled CH505 SOSIP design bound to the b12 Fab. (F) Left: Structure comparison between the closed-state b12-bound interprotomer disulfide-stapled CH505 SOSIP design and the open-occluded state b12-bound B41 isolate SOSIP. A single b12-bound gp120 domain from B41 is aligned to a closed-state CH505 design gp120 to highlight the shift in the b12 Fv position (only β sheets shown for clarity; pink arrows indicate shift direction). Two additional gp120 domains are shown as surfaces to highlight the potential for b12 clashes. Right: Alignment of the b12-bound apex-stapled CH505 SOSIP design and B41 isolate gp120 domains highlighting differences in the HCDR3 position.