Abstract
Antisense technology has great potential for the control of RNA expression, but there remain few successful applications of the technology. Expressed antisense RNA can effectively down-regulate expression of a gene over long periods, but cannot differentiate partly identical sequences, such as the mRNA of fusion genes or those with point mutants. We have designed a structured form of expressed antisense, which can discriminate between highly similar mRNA molecules. These ‘masked’ antisense RNAs have most of the antisense sequence sequestered within duplex elements, leaving a short single-stranded region to initiate binding to target RNA. After contacting the correct target, the structured RNA can unravel, releasing the masked antisense region to form a stable duplex with the mRNA. We demonstrate that suitable masked antisense RNA can discriminate between the two forms of BCR–ABL mRNA that result from the Philadelphia chromosomal translocations, as well as discriminating the normal BCR and ABL mRNA.
INTRODUCTION
The use of antisense to degrade or block the translation of mRNA has become an established technique to influence gene expression (Wagner and Flanagan, 1997), and short synthetic oligonucleotides are being extensively tested as potential therapeutic agents (Agrawal, 1996). However, where the presence of antisense is required over long periods, or within tightly defined tissue or cell types, expressed antisense RNA becomes a preferable option. If the target RNA is a unique gene product, e.g. viral genes in acute infection (Koschel et al., 1995; Offensperger et al., 1998) or ripening genes in fruit (Hamilton et al., 1995; Ayub et al., 1996), simply expressing the entire cDNA in the reverse orientation has been used with good effect. However, in other circumstances the desired target is not easily distinguishable from other RNAs, as in the case of mRNA from oncogenes with point mutations or from fusion genes resulting from tumour-specific chromosomal translocations. Often the normal proto-oncogenes are expressed alongside these aberrant transcripts. In such circumstances the application of expressed antisense is difficult as it cannot readily distinguish the target from its normal cellular counterpart(s).
Antisense-based gene controls occur naturally in prokaryotes, although the interaction of such antisense with their target mRNA is often a complex multi-step process, dependent on the three-dimensional structures of both antisense and sense RNA molecules (Wagner and Simons, 1994). While the structure of eukaryotic mRNAs cannot readily be determined, it is practical to incorporate secondary structure into an antisense RNA to modulate its interaction with potential target sequences (Patzel and Sczakiel, 1998). We have therefore designed structured antisense molecules in which the bulk of the antisense sequence is masked by partially complementary sequence in a hairpin structure and becomes available for hybridization only after a highly specific nucleation reaction with a short stretch of the RNA target. The validation of this approach has been undertaken using the BCR–ABL fusion mRNA, which is typical of those found in haematopoietic tumours and tumours of mesenchymal origin (Rabbitts, 1994).
The BCR–ABL fusion gene is associated with Philadelphia chromosome-positive chronic myelogenous leukaemia and acute lymphocytic leukaemia, resulting from t(9;22)(q34;q11) translocations. The fused genes encode proteins of 210 and 190 kDa, respectively. The difference between the BCR–ABL proteins reflects differences in the breakpoints within the BCR gene resulting in two mRNAs containing the same ABL exons fused to different BCR sequences (de Klein et al., 1982; Bartram et al., 1983; Heisterkamp et al., 1983; Groffen et al., 1984; Shtivelman et al., 1985; Hermans et al., 1987). Thus, the p210 and p190 BCR–ABL mRNA fusion junctions differ only on one side (i.e. the BCR side). In addition, the fusion mRNA is expressed alongside the normal BCR and ABL alleles in the leukaemic cells. We demonstrate that masked antisense RNA can successfully discriminate between p190 BCR–ABL, p210 BCR–ABL, BCR and ABL RNAs, providing an experimental framework for the design of highly discriminatory antisense reagents.
RESULTS AND DISCUSSION
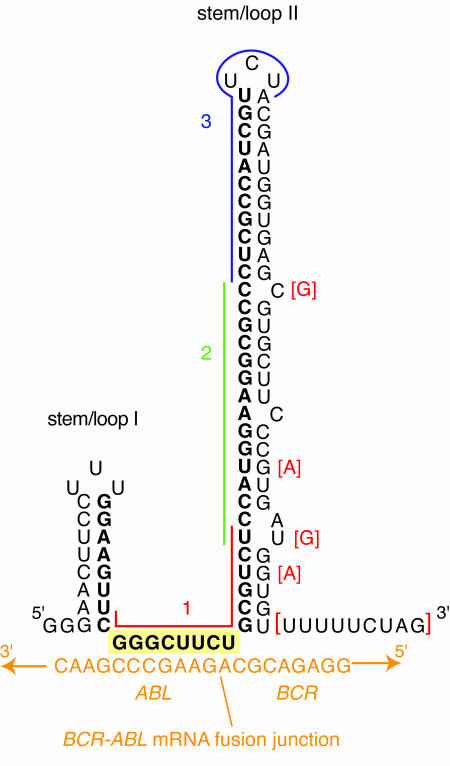
The masked antisense RNAs were designed as self-folding asymmetric dual hairpin structures (Figure 1) in which the bulk of the antisense sequence is hidden within the larger hairpin (stem–loop II). An eight nucleotide single-stranded region between the hairpins serves as the sequence that initially contacts the RNA target (Figure 1, yellow boxed sequence GGGCUUCU). In the example shown, hAS190 is a masked antisense against the p190 isoform of BCR–ABL. The hAS190 RNAs were designed around a 47 nucleotide stretch of sequence complementary to the fusion mRNA, with the capacity to self-fold being tested theoretically using the M-FOLD program (Jaeger et al., 1989a,b; Zuker, 1989) and confirmed experimentally by T1 RNase fingerprinting of in vitro transcripts (data not shown).
Fig. 1. Design of masked antisense molecules to target BCR–ABL mRNA. The sequence and putative secondary structure of masked antisense to BCR–ABL p190 (hAS190α) are shown. Antisense residues are shown in bold. The antisense molecule has two stem–loop structures of which stem–loop II contains 31 bases complementary to BCR. The targeting region (nucleation site) is highlighted in yellow and comprises eight bases complementary to the ABL sequence at the junction with BCR. The targeted region of BCR–ABL mRNA is shown in orange. The two forms of the hAS190 molecule, designated α and β, differ only in the descending strand of stem–loop II (the differences present in the hAS190β form are shown in red brackets). Coloured lines indicate the regions complementary to the oligonucleotides used for blocking experiments (see Figure 3); red, oligo 1; green, oligo 2; and blue, oligo 3.
The strategy was that the antisense would initially bind to its target (nucleation) via the single-stranded region in a transient manner, followed by progressive unwinding and hybridization if the target sequence is perfectly complementary to the antisense. Mismatched targets would either fail to form the initial transient interaction, or would not support the progression of hybridization into the hairpins. Stem–loop I seems to be required to initiate the desired folding of the molecule and can be formed from completely irrelevant sequence without affecting function (data not shown). Base mismatches and G:U pairs in the second hairpin are intended to lower the free energy of the folded antisense relative to the antisense/target hybrid, thereby influencing the rate of interaction of the two molecules. We generated two forms of hAS190 (α and β) to verify this, which differ at four residues in the descending arm of stem–loop II (Figure 1). We would expect equivalent mismatches to exert a greater influence when sited near the base of the stem–loop, although we have not tested this empirically.
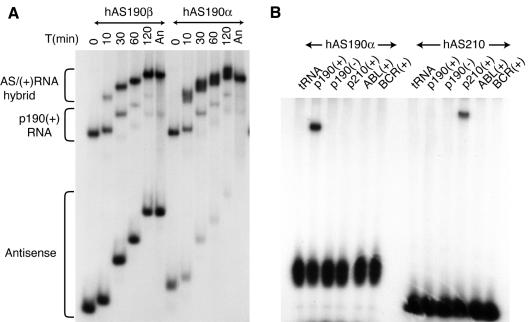
The two forms of hAS190 were tested for their ability to bind a target RNA generated by in vitro transcription from a plasmid containing a cDNA fragment of the p190 BCR–ABL gene. Target p190 RNA and antisense RNA were mixed in the presence of yeast tRNA and timed samples fractionated on continuously electrophoresing native polyacrylamide gels. Binding of p190 target by antisense resulted in the appearance of a third, slower migrating species in each lane (Figure 2A). The hybrid band was confirmed as a true hybrid between sense and antisense by performing interactions in which only the target or the antisense was radiolabelled (data not shown). The rate of hybrid formation appears to be inversely correlated with the calculated stability of the folded antisense. The α and β forms of folded hAS190 differ by12.4 kcal/mol in their free energies (hAS190α has a calculated free energy of –40.1 kcal/mol and hAS190β –52.5 kcal/mol). hAS190α was present in approximately equimolar amounts with p190 RNA, and complexed >50% of the target within 10 min (Figure 2A), whereas hAS190β complexed <20% of target RNA at 10 min despite being present in ∼5-fold molar excess.
Fig. 2. The interaction of hAS190 masked antisense RNA with the p190 RNA and related sequences. (A) Time course of interaction of hAS190 (α or β) with p190 BCR–ABL target RNA in vitro. Approximately 2 pmol of the p190 mRNA fragment [p190(+)RNA] were mixed with hAS190α (2 pmol) or hAS190β (10 pmol) antisense and incubated at 37°C. At the times indicated aliquots were removed and immediately loaded on a continuously electrophoresing 5% native polyacrylamide gel. Samples were run into the gel for 2 min at 10 V/cm then the voltage was reduced to 1 V/cm until the next sample was loaded. After the last sample was loaded, electrophoresis was completed at 10 V/cm. The bands indicated correspond to antisense–sense hybrid [AS/(+) RNA], unannealed p190 RNA [p190(+)] and antisense (either hAS190α or β).An, annealed RNAs. (B) Specificity of masked antisense interaction. Two picomoles of masked antisense to BCR–ABL p190 (hAS190α) or p210 (hAS210), labelled with 32P, were mixed with 0.2 pmol of the indicated unlabelled target mRNA species and allowed to associate at 37°C for 30 min, and the mixtures were resolved by gel electrophoresis, followed by autoradiography. Target mRNA fragments were: BCR–ABL fusion RNA, p190(+) or p210(+); normal ABL RNA, ABL(+); normal BCR RNA, BCR(+); and an antisense fragment of BCR–ABL p190, p190(–). tRNA indicates reaction carried out with only carrier tRNA.
The masked antisense and target RNA hybridize efficiently (Figure 2A). The ability of masked antisense RNA to discriminate between related RNA species (i.e. BCR–ABL p190 and p210, BCR and ABL) was investigated using hAS190α RNA and hAS210 (designed to bind the p210 BCR–ABL form). Various in vitro target RNA transcripts were incubated with excess radiolabelled hAS190α or hAS210, followed by native gel electrophoresis. hAS190α could only form a hybrid with the p190 mRNA strand [Figure 2B, p190(+)] and not with BCR, ABL, p210 BCR–ABL or the negative strand of p190 BCR–ABL [p190(–)], despite these RNAs containing sequence elements complementary to the antisense. Conversely, the antisense molecule hAS210 bound only the p210 RNA [Figure 2B, p210(+)]. These results demonstrate that the masked antisense molecules can efficiently and specifically bind a target in vitro. Confirmation that the unwinding and re-hybridization of the hairpins goes to completion was obtained by RNase fingerprinting of the hybrids, which yielded the same T1 RNase resistant fragments as did the conventional hybridization of the two species by heat denaturation and slow re-annealing (data not shown).
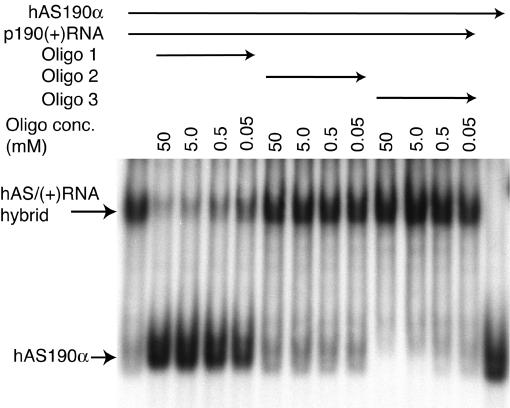
Complete hybrid formation of sense RNA with the masked antisense occurs in very mild conditions of temperature (37°C) and salt, and does not require prior denaturation of the antisense. This suggested that the folded antisense RNA unwinds after an initial nucleation contact on its specific target and that binding to the sense RNA through the targeting region is essential for hybrid formation. This was confirmed by blocking experiments with short oligonucleotides complementary to three regions of the hAS190α antisense molecule (Figure 1). Radiolabelled hAS190α RNA was incubated briefly with increasing amounts of oligonucleotide prior to the addition of p190 target RNA [p190(+) RNA]. Native gel electrophoresis was used to assess the effect on hybrid formation (Figure 3). No competition effect was found with oligonucleotides 2 or 3, which are complementary to the ascending strand of stem–loop II and the apical stem–loop region, respectively. However, oligonucleotide 1, which is complementary to the nucleation sequence, was effective in blocking the hybrid formation. Thus, the initial hybridization most likely only occurs via the targeting region and propagates into the hairpins.
Fig. 3. Blocking of the masked antisense–sense RNA interaction by oligonucleotide competition. 32P-labelled hAS190α (1 pmol) was mixed with the indicated concentrations of unlabelled oligonucleotide and incubated at room temperature for 2 min. Unlabelled mRNA fragment p190(+) RNA (1 pmol) was added and the reaction incubated at 37°C for 30 min before samples were resolved by gel electrophoresis, followed by autoradiography. The competing oligonucleotides were complementary to the three regions indicated in Figure 1.
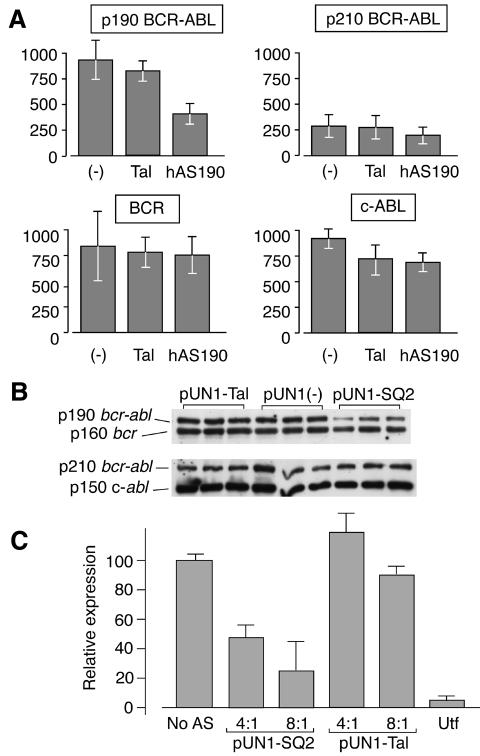
As hybrid formation occurs between folded antisense and its target without denaturation (Figure 2), these molecules should be effective in vivo. Therefore, structured antisense RNAs were expressed in vivo using an expression vector (pUN1) based on the human U6 snRNA promoter. This vector is capable of generating transcripts that contain no vector-derived sequence other than the essential G at position +1 and a U4 tail. Since BCR–ABL proteins (and BCR and ABL) seem to have long half-lives in vivo (Dhut et al., 1990; Spiller et al., 1998), we performed efficacy studies in HeLa cells by co-transfecting pUN1-based antisense constructs with p190 or p210 BCR–ABL expression vectors, and quantitating the levels of BCR–ABL protein after 16 h. Figure 4A shows the collated results from three sets of triplicate assays. Co-transfection of the pUN1.hAS190 construct with a p190 BCR–ABL expression clone resulted in a >50% reduction in the expression of p190 BCR–ABL compared with a non-relevant antisense control (pUN1.Tal) or with the empty pUN1 vector (Figure 4A). When p190 masked antisense was co-transfected with a p210 expression clone, the levels of p210 BCR–ABL were slightly lower than in the presence of Tal1 antisense, but not significantly, as judged by triplicate experiments (Figure 4A). The levels of endogenous Bcr or Abl were unaffected comparing transfection of p190 and Tal1 antisense, as judged by western analysis (Figure 4B). We also found that the effect of hAS190 was dependent on the ratio of specific antisense to the BCR–ABL expression vector, with increasing effect as the ratio of antisense increased (Figure 4C). No such effect was found with the non-specific antisense molecule, pUN1-Tal (Figure 4C), although promoter interference was problematic in the context of this assay. This was particularly noticeable when using standard expressed antisense, driven from polII promoters, for comparison with the masked antisense. The maximum effect attributable solely to standard expressed antisense was a reduction of ∼70% in p190 expression (data not shown). Thus, the efficacy and specificity of the hAS190 RNA are largely retained in vivo. These data indicate that use of the polIII promoter system produces an RNA product that, like the in vitro product, can self-fold and specifically interact with RNA targets.
Fig. 4. In vivo effect of expressed masked antisense on BCR–ABL production. (A) HeLa cells were co-transfected with BCR–ABL expression vectors and antisense-expressing polymerase III vectors (see Methods). After 16 h, levels of BCR–ABL, ABL and BCR proteins were assessed by western blotting with specific antibodies. Values are the integrated areas of specific bands measured by scanning densitometry and represent the average, plus standard deviation, of nine independent transfections. The levels of p190 BCR–ABL, p210 BCR–ABL and endogenous BCR or ABL are shown. Each histogram is for co-transfection of reporter plasmid with empty vector (–), non-relevant Tal1 antisense (Tal) or masked p190 BCR–ABL antisense (hAS190). (B) Representative western blots showing triplicate transfections of p190 (upper panel) or p210 (lower panel) BCR–ABL reporter plasmids with either pUN1-Tal1, pUN1(–) or pUN1-SQ2 antisense constructs. The blots were probed with anti-BCR (upper panel) or anti-ABL (lower panel) antisera. (C) Titration of antisense versus reporter plasmids. The p190 expression vector was transfected into HeLa cells alone (No AS) or together with the indicated molar ratios of pUN1-SQ2 or pUN1-Tal antisense vector. Expression was measured as in (A). Values are the average of two independent transfections. Utf, untransfected HeLa cells.
Our data demonstrate that the behaviour of an RNA, in particular its capacity to form a duplex with a complementary sequence, can be modulated by the incorporation of secondary structure. By masking the majority of an antisense sequence within stem–loops, we have reduced the region of sequence that is available to initiate contact between the antisense and its target to a few nucleotides. This contact is insufficient to produce a stable interaction. The formation of a stable hybrid is dependent upon the progression of the antisense binding into the second hairpin, which requires that the target sequence is perfectly complementary to the antisense along its entire length. We have made masked antisense for a fusion mRNA (BCR–ABL) by positioning the structural elements of the antisense so that the initial contact occurs on one side of the fusion junction (ABL sequences) while the bulk of the antisense is complementary to the BCR sequences on the other side of the junction. After initial binding to the ABL sequences, breathing of the stem–loop II duplex allows further interaction with increasing lengths of the BCR sequences, resulting in a progressive unwinding in stem–loop II with the simultaneous hybridization of the antisense along the fusion mRNA. This results in a highly stable duplex. Binding to the other BCR–ABL (p210) isoform or normal ABL is precluded as the complementarity of target and antisense does not extend into stem–loop II. Similarly, binding to normal BCR is precluded as no complementary sequence is available in the folded antisense to make the initial contact. The result is a reagent that is specific for the p190 fusion mRNA.
The generation of structured antisense molecules has the potential to bind specifically to and down-regulate one member of a group of highly similar RNA targets. The de novo design of similar antisense molecules against other target RNA species should be relatively simple based upon the general structure of the hAS RNAs, or any subsequently improved structure, with the aid of the M-FOLD program. It is likely that by alteration of the length of the targeting region, the position of its binding to target RNA and the stability of the masked hairpin we should be able to discriminate even point mutations using the masked antisense system. Further refinement of the optimal configurations should derive consensus structures on which the rational design of any desired antisense can be based.
METHODS
Vectors and RNA synthesis. Antisense RNA templates were synthesized from oligonucleotides and cloned into the vector pT7c. Antisense RNAs were transcribed from in vitro transcription reagents (Ambion) including [32P]UTP (600 Ci/mmol; Amersham) as required. Target RNAs were transcribed from the T7 or T3 promoters of pBluescriptII, the templates being fragments of the target gene cDNAs (p190 BCR–ABL, BglII–KpnI; p210 BCR–ABL, HindIII–KpnI; c-ABL, NarI–KpnI; and BCR, BamHI–SalI). All construct sequences were verified.
The sequences of hAS190α and β are shown in Figure 1. hAS210 has the sequence: 5′-GGGCGAAUUGGAUUCGCCCGGGCUUUUGAACUCUGCUUAAAUCCAGUGGCUGAGUGGAUCUUCCACUUACUACUGGACUUAAGUAGUGUUCAUGCAUCUAG-3′.
Interaction of masked antisense RNAs with target RNAs in vitro. RNA interactions were carried out at 37°C in 250 mM NaCl, 10 mM Tris–HCl pH 8 containing 0.5 mg/ml yeast tRNA. Reaction volumes were 20 µl except for the timed series (Figure 2A) where 5 µl aliquots were taken at the times indicated from a 40 µl reaction. Reactions were stopped by mixing with an equal volume of ice-cold glycerol buffer (40% glycerol, 20 mM Tris–HCl pH 8, 10 mM EDTA) or loading buffer (95% formamide, 5 mM EDTA pH 8), for native and denaturing polyacrylamide gels, respectively.
Oligonucleotide competition. Approximately 1 pmol of 32P-labelled hAS190a was premixed with oligonucleotide in reaction buffer and incubated at RT for 2 min. Unlabelled p190 mRNA fragment (1 pmol) was added and the reaction incubated at 37°C for 30 min. The reactions were run on a 5% native polyacrylamide gel prior to autoradiography. Blocking oligonucleotides were 15- or 16mers with similar predicted Tm values (48–50°C). The sequences are: oligo 1, 5′-AGACGCAGAAGCCCG; oligo 2, 5′-GTAGAACGATGGCGAG; oligo 3, 5′-GGCGCCTTCCATGGA.
Expression vectors and HeLa cell transfections. Expression vectors containing p190 and p210 BCR–ABL c-DNAs (pE1A2 and pKW3, respectively) were a gift from Professor G. Grosveld. Antisense expression constructs were made in the vector pUN1, which contains a polIII promoter from the human U6 snRNA gene.
HeLa cells were seeded into 6-well plates at 1–3 × 106 cells/well and grown overnight. Transfection was carried out with a constant amount of DNA (4 µg) and Superfect (Qiagen) (10 µl) per well. For each co-transfection the mass amounts of reporter plasmid were kept constant and antisense vectors were mixed with pGEM3zf(+) plasmid to give a constant molar ratio of reporter to antisense as well as a constant total mass of DNA. Sixteen hours after transfection, the cells were harvested and lysed by boiling in SDS sample buffer. Samples were resolved on 5–15% SDS–PAGE gels, and electroblotted onto PVDF membranes. BCR, ABL and BCR–ABL proteins were detected using either a rabbit anti-Bcr antiserum (Santa Cruz Biotechnology Inc.) or a monoclonal anti-Abl (8E9), followed by an HRP-conjugated secondary antibody and ECL reagents (Amersham). The resulting film images were quantified using a scanning densitometer. Transfection efficiencies were routinely 30–40% as determined by flow cytometry using 0.1 µg of pEYFP-C1 (Clontech) added into transfection mixtures.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Medical Research Council. We would like to thank Alan Forster for expert help during this work. We also thank Professor G. Grosveld for the BCR–ABL cDNA clones and Dr J. Wang for the 8E9 anti-Abl antibody.
REFERENCES
- Agrawal S. (1996) Antisense oligonucleotides: towards clinical trials. Trends Biotechnol., 14, 376–387. [DOI] [PubMed] [Google Scholar]
- Ayub R., Guis, M., Amor, M.B., Gillot, L., Roustan, J.-P., Latche, A., Bouzayen, M. and Pech, J.-C. (1996) Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nature Biotechnol., 14, 862–866. [DOI] [PubMed] [Google Scholar]
- Bartram C.R. et al. (1983) Translocation of c-abl oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature, 306, 277–280. [DOI] [PubMed] [Google Scholar]
- de Klein A. et al. (1982) A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature, 300, 765–767. [DOI] [PubMed] [Google Scholar]
- Dhut S., Chaplin, T. and Young, B.D. (1990) BCR–ABL and BCR proteins: biochemical characterization and localization. Leukemia, 4, 745–750. [PubMed] [Google Scholar]
- Groffen J., Stephenson, J.R., Heisterkamp, N., de Klein, A., Bartram, C.R. and Grosveld, G. (1984) Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromsome 22. Cell, 36, 93–99. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J., Fray, R.G. and Grierson, D. (1995) Sense and antisense inactivation of fruit ripening genes in tomato. Curr. Top. Microbiol. Immunol., 197, 77–89. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N., Stephenson, J.R., Groffen, J., Hansen, P.F., de Klein, A., Bartram, C.R. and Grosveld, G. (1983) Localization of the c-abl oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature, 306, 239–242. [DOI] [PubMed] [Google Scholar]
- Hermans A. et al. (1987) Unique fusion of bcr and c-abl genes in Philadelphia chromosome positive acute lymphoblastic leukemia. Cell, 51, 33–40. [DOI] [PubMed] [Google Scholar]
- Jaeger J.A., Turner, D.H. and Zuker, M. (1989a) Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA, 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J.A., Turner, D.H. and Zuker, M. (1989b) Predicting optimal and suboptimal secondary structure for RNA. Molecular Evolution: Computer Analysis of Protein and Nucleic Acid Sequence. pp. 281–306. [DOI] [PubMed]
- Koschel K., Brinckmann, U. and Hoyningen-Huene, V.V. (1995) Measles virus antisense sequences specifically cure cells persistently infected with measles virus. Virology, 207, 168–178. [DOI] [PubMed] [Google Scholar]
- Offensperger W.B., Offensperger, S. and Blum, H.E. (1998) Antisense therapy of hepatitis B virus infection. Mol. Biotechnol., 9, 161–170. [DOI] [PubMed] [Google Scholar]
- Patzel V. and Sczakiel, G. (1998) Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nature Biotechnol., 16, 64–68. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. (1994) Chromosomal translocations in human cancer. Nature, 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Shtivelman E., Lifshitz, B., Gale, R.P. and Canaani, E. (1985) Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature, 315, 550–554. [DOI] [PubMed] [Google Scholar]
- Spiller D.G., Giles, R.V., Broughton, C.M., Grzybowski, J., Ruddel, C.J., Tidd, D.M. and Clark, R.E. (1998) The influence of target protein half-life on the effectiveness of antisense oligonucleotide analog-mediated biologic responses. Antisense Nucleic Acid Drug Dev., 8, 281–293. [DOI] [PubMed] [Google Scholar]
- Wagner E.G.H. and Simons, R.W. (1994) Antisense RNA control in bacteria, phages and plasmids. Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- Wagner R.W. and Flanagan, W.M. (1997) Antisense technology and prospects for therapy of viral infections and cancer. Mol. Med. Today, 3, 31–38. [DOI] [PubMed] [Google Scholar]
- Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]