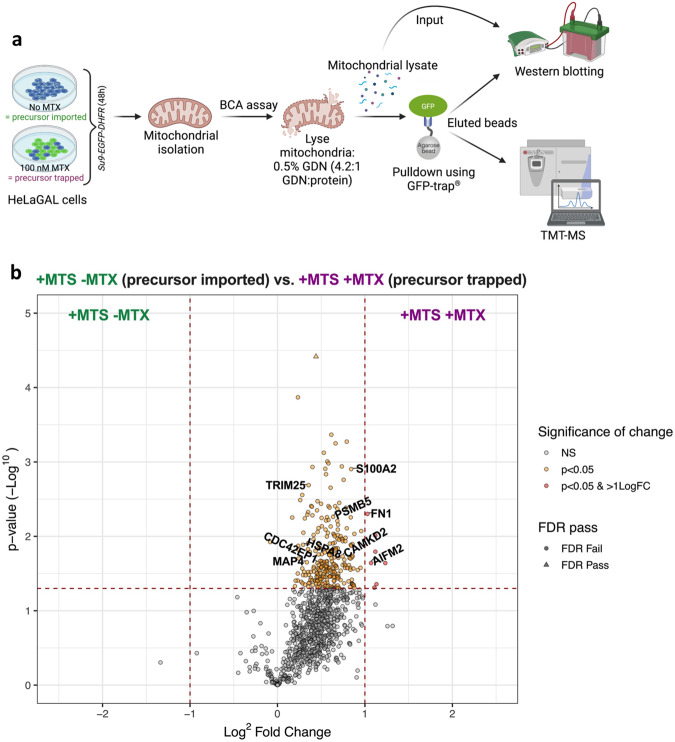
Fig. 7. TMT-MS highlights TNT-associated proteins enriched with trapped precursor.
a Schematic showing TMT-MS experimental outline. HeLaGAL cells were subjected to Su9-EGFP-DHFR over-production in the presence or absence of 100 nM MTX for 48 h. Mitochondria were isolated and their protein content was assessed using a BCA assay. Equal amounts of mitochondria were lysed using 0.5% GDN at a ratio of 4.2:1 (GDN: protein by mass). A fraction of mitochondrial lysate was saved as an input control, and the rest was loaded onto GFP trap agarose beads, to pulldown proteins associated with the GFP-tagged protein of interest (trapped or imported precursor). The resulting beads conjugated to proteins associated with GFP-tagged proteins were analysed by Western blot and TMT-MS. Schematic created using BioRender. b Volcano plot highlighting proteins enhanced in pulldown samples from mitochondria with imported precursor (+ MTS - MTX, left) or trapped precursor (+ MTS + MTX, right). Proteins of interest are highlighted by gene names. N = 3 biological replicates. Statistical significance was determined using a two-tailed, unpaired t test (b). Raw data are provided in the Supplementary Data file (b). BCA bicinchoninic acid, FDR false discovery rate, GDN glycol-diosgenin, LogFC Log2 fold change, MTS mitochondrial targeting sequence, MTX methotrexate, NS not significant, TMT-MS tandem mass tagging mass spectrometry, TNT tunnelling nanotube.