Key Points
-
•
Comprehensive genomic profile of patients with FPDMM with germ line RUNX1 mutations.
-
•
Rising clonal hematopoiesis related secondary mutations that may lead to myeloid malignancies.
Visual Abstract
Abstract
Familial platelet disorder with associated myeloid malignancies (FPDMM) is caused by germline RUNX1 mutations and characterized by thrombocytopenia and increased risk of hematologic malignancies. We recently launched a longitudinal natural history study for patients with FPDMM. Among 27 families with research genomic data by the end of 2021, 26 different germline RUNX1 variants were detected. Besides missense mutations enriched in Runt homology domain and loss-of-function mutations distributed throughout the gene, splice-region mutations and large deletions were detected in 6 and 7 families, respectively. In 25 of 51 (49%) patients without hematologic malignancy, somatic mutations were detected in at least 1 of the clonal hematopoiesis of indeterminate potential (CHIP) genes or acute myeloid leukemia (AML) driver genes. BCOR was the most frequently mutated gene (in 9 patients), and multiple BCOR mutations were identified in 4 patients. Mutations in 6 other CHIP- or AML-driver genes (TET2, DNMT3A, KRAS, LRP1B, IDH1, and KMT2C) were also found in ≥2 patients without hematologic malignancy. Moreover, 3 unrelated patients (1 with myeloid malignancy) carried somatic mutations in NFE2, which regulates erythroid and megakaryocytic differentiation. Sequential sequencing data from 19 patients demonstrated dynamic changes of somatic mutations over time, and stable clones were more frequently found in older adult patients. In summary, there are diverse types of germline RUNX1 mutations and high frequency of somatic mutations related to clonal hematopoiesis in patients with FPDMM. Monitoring changes in somatic mutations and clinical manifestations prospectively may reveal mechanisms for malignant progression and inform clinical management. This trial was registered at www.clinicaltrials.gov as #NCT03854318.
Introduction
RUNX1 is a transcription factor indispensable for the development and function of definitive hematopoietic stem cells (HSCs).1 Chromosome translocations and somatic mutations affecting RUNX1 are frequently detected in hematologic malignancies, such as myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and acute lymphoblastic leukemia (ALL).2 Germline RUNX1 mutations lead to familial platelet disorder with associated myeloid malignancy (FPDMM; Online Mendelian Inheritance in Man no. 601399), a rare autosomal dominant disease associated with platelet defects, both quantitatively and qualitatively, resulting in easy bleeding and bruising.3 Patients with FPDMM are predisposed to hematologic malignancies,4,5 for example, MDS, AML, chronic myelomonocytic leukemia, or ALL.6, 7, 8 The current consensus for the incomplete penetrance of malignancy is that germline RUNX1 mutations are insufficient for leukemogenesis. Additional risk factors, such as somatic mutations, are important for the development of hematologic malignancies.4,9
Somatic mutations in HSCs may lead to accelerated proliferation and reduced cell death, resulting in clonal expansion of the mutation-carrying HSC, or clonal hematopoiesis (CH).10,11 CH increases with age. Large population studies showed that CH increases the risks of atherosclerotic cardiovascular diseases,12,13 hematologic neoplasms,11,14 and other nonmalignant diseases.15 Previous studies have described early onset of CH in patients with FPDMM without hematologic malignancy.4,16 However, it is still unclear what the role of CH is in the development of hematologic malignancies.
To improve our understanding of FPDMM pathogenesis and identify potential driver alterations for malignancy transformation, we initiated a natural history study in 2019 to longitudinally investigate the genomic and clinical profile of FPDMM. Here, we report the genomic data from 62 patients enrolled in our study whose samples had been sequenced for research purposes by the end of 2021.
Methods
Patients and samples
Patients were enrolled in the clinical study entitled “Longitudinal Studies of Patients with FPDMM,” after obtaining informed consent in accordance with the declaration of Helsinki. RUNX1 variants were determined to be pathogenic (P), likely pathogenic (LP), or variants of uncertain significance (VUSs) by American College of Medical Genetics (ACMG) ClinGen Myeloid Malignancy Variant Curation Expert Panel criteria.17 Clinical studies of the enrolled participants have been described recently by Cunningham et al.18
Genomic DNA, RNA, and cryopreserved cell samples were processed and biobanked for further needs.
Exome sequencing and data processing
Exome sequencing (ES) of genomic DNA samples at the NIH Intramural Sequencing Center (NISC) is described in supplemental Methods. In brief, genomic DNA were fragmented to the size around 300bp, ligated and pre-amplified with adapters, following exome panels capture. Libraries were sequenced on NovaSeq 6000 platform with PE151 strategy. Our IDT xGen Exome Research Panel data achieved a mean coverage of 87X-353X, with a median of 174X, expect to identify somatic mutations with variant allele frequency (VAF) above 3-5% in most of the region, and lower VAF at regions with higher coverage. Detailed information about the sequenced samples is described in supplemental Table 1 and supplemental Figure 1. Sequencing data were analyzed with in-house pipelines on NIH high-performance computing system “Biowulf.” Detailed descriptions of data analysis, workflow and parameters can be found in supplemental Figure 2 and supplemental Methods. All RUNX1 mutations listed in this manuscript are based on the representative transcript NM_001754 (coding RUNX1c).19
Bulk RNA-seq and analysis
RNA sequencing (RNA-seq) was performed at NISC with Illumina TruSeq stranded chemistry and PE151 strategy on NovaSeq 6000. The sequenced samples are listed in supplemental Table 1, and detailed information on sequencing and data analysis is described in supplemental Methods. Published healthy donor bone marrow (BM) RNA-seq data were used for splice junction analysis.20
Cytogenetics and CNV analyses
Cytogenetic analyses of BM cells were conducted at the Mayo Clinic Laboratories. For single-nucleotide polymorphism (SNP) array, Infinium OmniExpressExome-8 kit was used to analyze guide DNA samples (supplemental Table 1). CNVPartition and PennCNV21 were used to identify candidate copy number variation (CNV) events. CNVkit22 (version 0.9.8) was used to identify CNVs from ES data for samples without SNP-array data. All CNV calls were revised with Integrative Genomics Viewer illustration.
Results
Study cohort and RUNX1 variant evaluation
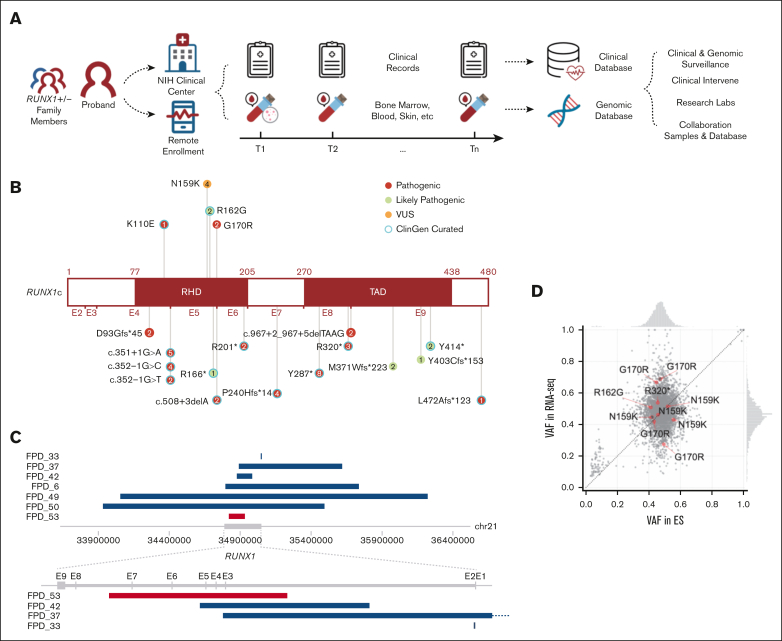
The natural history study was launched in early 2019, and by the end of 2021, 111 patients and 45 family controls had been enrolled. This report focuses on 62 patients in 27 families whose research genomic data were available. Sequential samples had been collected for 19 patients who visited NIH Clinical Center (NIHCC) more than once (Figure 1A; supplemental Table 1).
Figure 1.
Overview of NIH FPDMM natural history study. (A) Study schema. Enrolled patients with germline RUNX1 variants and family member controls visited the National Institutes of Health (NIH) clinical center and/or sent remote samples annually. Both clinical and research data stored in the clinical and genomic databases were used for downstream research and guide clinical management. (B) RUNX1 mutations carried by the patients are included in this manuscript. Numbers in the circle indicate patient number who carries the mutation. (C) CNVs affecting RUNX1 gene. Blue bars are showing deletions, and red bars are showing duplication. (D) VAF correlation between exome sequencing and RNA-seq data. Red dots are RUNX1 variants, and gray dots are all other variants (all variants with ≥20× coverage in both RNA-seq and ES, including both germline and somatic) in all samples that have 2 platform data set. Histograms (top and right) show the VAF distributions in exome sequencing and RNA-seq, respectively.
Our cohort excluded families carrying benign, likely benign, or VUSs without enough evidence of familial platelet disorder (FPD)-like clinical features. In total, 26 different germline RUNX1 variants were detected in the 27 families. The most common types of RUNX1 mutations are mutations causing truncated RUNX1 protein (including splice-site mutations, frameshift mutations, and stop-gain mutations), or large CNVs that cause complete loss or partial loss or gain of the RUNX1 gene (Figure 1B-C; supplemental Table 2). All RUNX1 mutations listed here are based on NM_00175419). Four families had 4 different missense variants; all located in the Runt homology domain, with 3 predicted to be P or LP, and 1 predicted to be VUS per ACMG ClinGen Myeloid Malignancy Variant Curation Expert Panel criteria.17 We included the family (FPD_5) with a RUNX1 VUS variant (c.477T>G, p.Asn159Lys) in the study because all 5 RUNX1 variant carriers in this family (across 3 generations) had mild-to-moderate thrombocytopenia as well as abnormal platelet functions and/or platelet morphological abnormalities. On the other hand, 2 noncarriers from the family, who were tested, had normal platelet counts.
Large-scale genomic alteration is common in our study cohort. Among the 27 families, 3 had large deletions covering the entire RUNX1 gene and, in some cases, additional genes. Three other families had smaller deletions, and 1 family has an intragenic duplication that affects several RUNX1 exons (Figure 1C).
To determine whether the mutations alter RUNX1 expression, we compared allele frequencies between ES and RNA-seq data from 7 patients, which have adequate coverage (including both germline and somatic mutations). The RUNX1 variant alleles were expressed between 40% and 70% at the RNA level, in the expected germline variant allele frequency (VAF) range (Figure 1D).
RUNX1 splice-site mutations
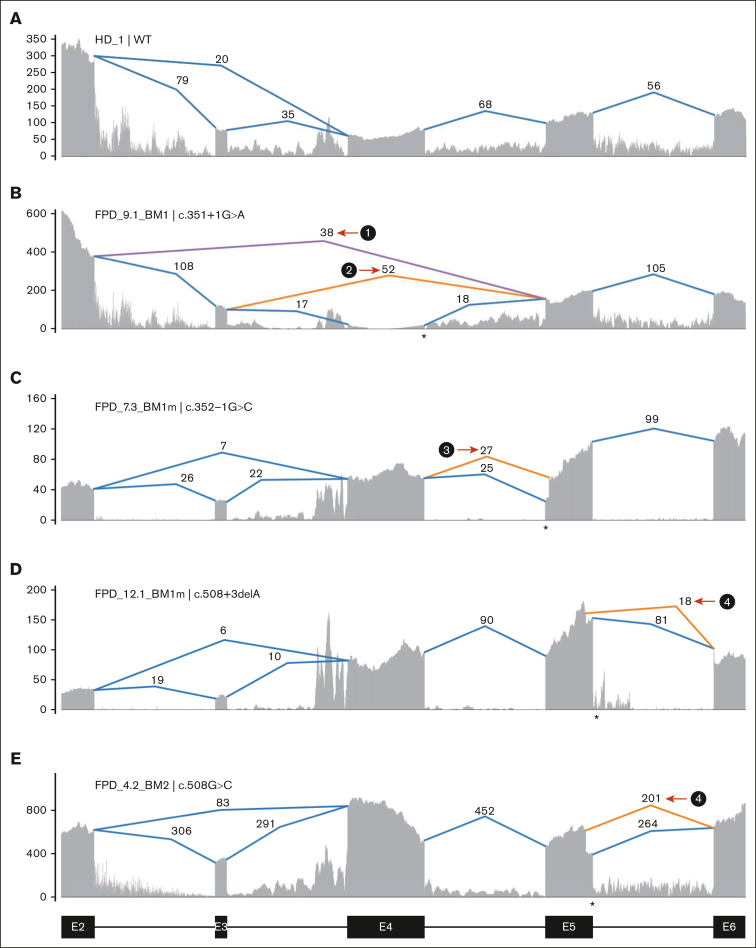
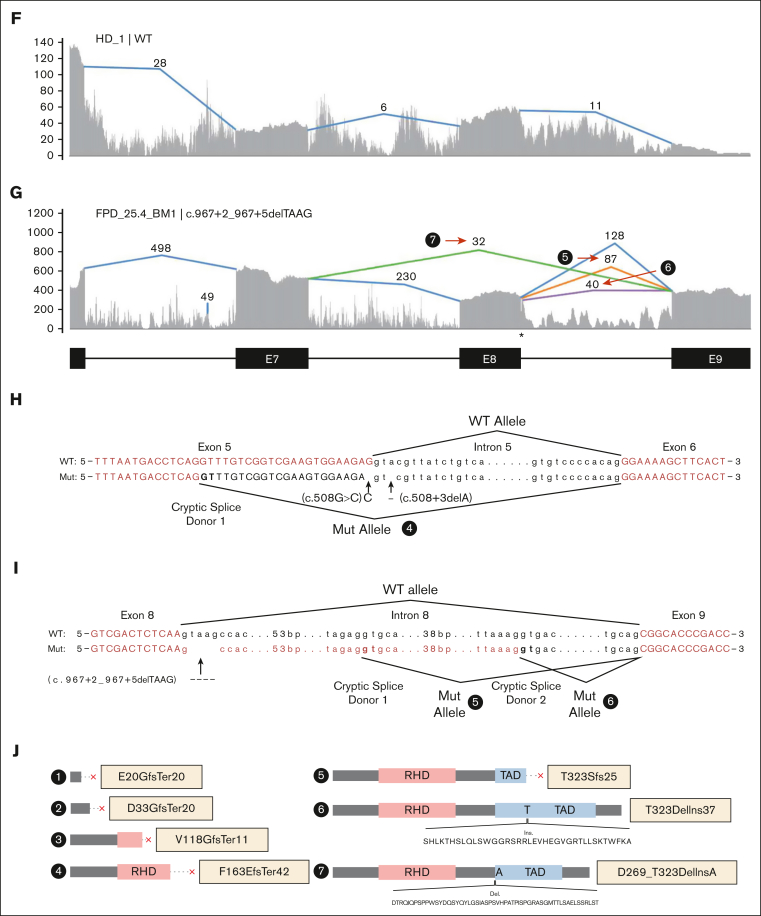
Multiple families have mutations at or near splice donor or acceptor sites, located in introns 4, 5, and 8 (Figure 1B). These mutations led to aberrantly spliced transcripts, as detected by RNA-seq (Figure 2; supplemental Figure 3). The c.351+1G>A variant, detected in 2 independent families, caused 2 types of exon-4 skipping: E2 to E5 and E3 to E5 (Figure 2B; supplemental Figure 3B). Junction counts showed a significantly higher proportion of the novel splicing products than the wild-type splicing products. The c.352-1G>C variant led to the usage of a cryptic acceptor site in exon 5 (Figure 2C; supplemental Figure 3C), which is also the cryptic acceptor site resulting from a c.352-1G>T variant.3 In this case, the mutant and wild-type transcripts were detected at similar levels. A cryptic splice donor site near the end of exon 5 was identified in patients with c.508+3delA (Figure 2D; supplemental Figure 3D), as previously reported.23 Interestingly, a missense variant c.508G>C (p.G170R) affects the adjacent splice donor site at the end of exon 5, leading to the usage of the same cryptic splice donor site associated with c.508+3delA (Figure 2E,H; supplemental Figure 3D); only 5% to 10% of transcripts are in the missense form. For both c.508+3delA and c.508G>C variants, the splice product resulting from the cryptic donor site had fewer aberrant splicing junction counts when than the wild-type junction, but there is a strong signal of intron retention (Figure 2D-E; supplemental Figure 3D). Finally, a c.967+2_967+5delTAAG variant caused exon 8 skipping (Figure 2G,I; type 7) and the activation of cryptic splice donors in intron 8 (Figure 2G,I; types 5 and 6); 1 of them (type 5) has been reported previously.24 At the protein level, it is predicted that the first 5 splice-site–related variants will produce truncated RUNX1 proteins, whereas the c.967+2_967+5delTAAG variant will lead to 2 in-frame products: an insertion of 37 amino acids in the middle of transactivation domain (TAD) domain for type 6 and a deletion of 55 amino acids at the beginning of the TAD domain for type 7 (Figure 2J). Most of the splice-site variants reported lead to an early stop codon, similar to nonsense or frameshift variants; we did not observe a reduced transcript expression level from the RUNX1 mutant allele caused by nonsense-mediated messenger RNA decay.25 Most of the RUNX1 splice-site variants had an estimated VAF of ∼50% in RNA-seq, but the c.351+1G>A samples even showed higher mutant allele VAF of ∼84% (supplemental Table 1).
Figure 2.
Aberrant splicing events in patients with RUNX1 splice-site variants. (A) Splice junctions and covered reads from exons 2 to 6 of RUNX1 in a heathy donor BM sample. (B-E) Splice junctions and covered reads in 4 patients with different RUNX1 splice-site or splice-region mutations. Orange or purple lines indicate abnormal junctions and blue lines indicate junctions also seen in the healthy control. Numbers indicate supporting read counts. (F) Splice junctions and covered reads from exons 6 to 9, in a heathy donor BM sample. (G) Splice junctions and covered reads in a patient with c.967+2_967+5delTAAG mutation. (H-I) Detailed information of cryptic splice donors and abnormal splice junctions caused by c.508+3delA, c.508G>C, and c.967+2_967+5delTAAG. (J) Predicted protein products of the 6 abnormal splicing transcripts shown in panels B-E and G. HD, healthy donor; Mut, mutation; WT, wild-type.
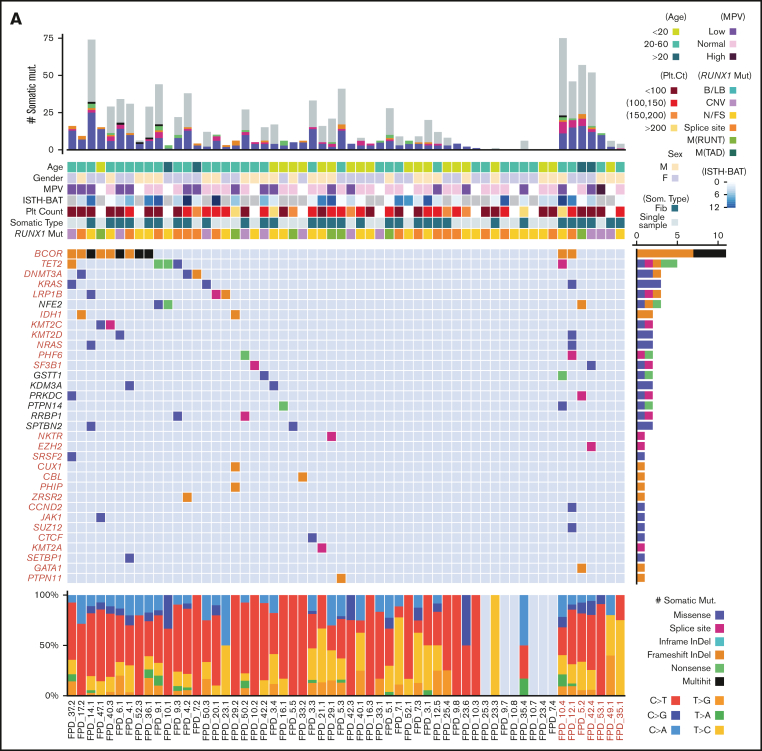
Somatic mutation landscape in patients with FPDMM
We have generated ES data from 58 patients with FPDMM for somatic mutation identification. We applied 2 strategies (supplemental Figure 2) to identify somatic mutations. For 31 patients with ES data from fibroblast, true somatic mutations were confirmed by comparing BM/peripheral blood (PB) data with fibroblast data. For the remaining 27 patients without fibroblast data, we used Mutect2 single sample mode to identify likely somatic mutations in the PB or BM samples with a panel of normal reference, which was composed of sequencing data from all unaffected family members in our cohort. The variants were further verified according to their population frequency (at <1%), absence in any members of the same family, and presence in the Catalogue of Somatic Mutations in Cancer database. The number of identified somatic mutations in these patients is likely an underestimate because we have been conservative with somatic mutation calling.
The somatic mutation landscape of hematopoietic cells in the patients with FPDMM of our cohort is depicted in Figure 3A. The middle heat map shows the aggregated somatic mutation landscape that merged all mutations in CHIP- or AML-driver genes26,27 (CL genes for CHIP- and leukemia-driver genes are listed in supplemental Table 3) detected in each patient and recurrently detected in multiple individuals. Interestingly, 25 of 51 (49%) patients without hematologic malignancy and 4 of 7 (57%) patients with hematologic malignancy have at least 1 somatic mutation in CL genes (supplemental Table 3). Somatic mutations were recurrently (>1 patient) observed in the following CL genes: BCOR, TET2, DNMT3A, KRAS, LRP1B, IDH1, KMT2C, KMT2D, NRAS, PHF6, and SF3B1. BCOR was the most frequently mutated CL gene because BCOR mutations were found in 11 of 58 patients (19%) and most BCOR mutations resulted in frameshifts. Moreover, 4 patients had >1 somatic BCOR mutation detected at the same time. Recurrent mutations were also obsereved in 7 non-CL genes (NFE2, GSTT1, KDM3A, PRKDC, PTPN14, RRBP1, and SPTBN2).
Figure 3.
Somatic mutations of the NIH FPDMM cohort. (A) Somatic mutation landscape depicting CHIP genes, AML-driver genes, and recurrently mutated genes. Each column is a patient; patients with hematologic malignancy are highlighted in red. The center heat map shows somatic mutation distribution; mutation types are represented with specific colors, as described by legends at the bottom right. Each row is a gene; genes belonging to CHIP and AML-driver genes are highlighted in red. The next heat map (top) shows annotated demographic and clinical information, which are also color-coded, as described by the legends at the right-hand side. The top bar plot shows the total somatic mutation numbers in each patient, with coding region mutations marked with colors used in the middle heat map and the noncoding region mutations colored in gray. The bar plot on the right of the middle heat map shows aggregated somatic mutation numbers for each gene, with color coding for mutation types. The bottom bar plot shows the percentage of the different base-changes (C>T, C>G, etc) in each patient. (B) Correlation between patients’ age at the time of sampling and the total number of somatic mutations. Only samples with fibroblast controls are included. Each dot in the graph represents the number of somatic mutations in 1 sequenced sample from a patient at the indicated age, and the color indicates the type of RUNX1 germline mutation in that patient. (C) Functional enrichment analysis of all somatically mutated genes. B/LB, benign/likely benign; CNV, copy number variation; fib, fibroblast; InDel, insertion-deletion; ISTH-BAT, International Society on Thrombosis and Haemostasis Bleeding Assessment Tool; MPV, mean platelet volume; M(RUNT), mutation in RUNT domain; M(TAD), mutation in transactivation domain; N/FS, nonsense/frame shift; Plt, platelet; som, somatic.
The overall somatic mutation numbers in each patient are shown in the top bar graph in Figure 3A. There seemed to be a correlation between the overall number of somatic mutations and the presence of CL gene mutations. Notably, 26 of 51 (51%) patients without a hematologic malignancy had no mutation detected in CL genes. However, somatic CL mutations in patients without fibroblast ES data might have been underdetected (CL mutations were found in 14 of 25 patients with fibroblast ES data, whereas in only 11 of 26 patients without such data). As expected, the total numbers of somatic mutations correlated with patients’ ages (Figure 3B; supplemental Figures 4B and 8B). Based on the published data from The Cancer Genome Atlas (TCGA) program, myeloid malignancies showed a relatively lower mutation burden than other cancer types.28,29 For the 43 patients without a hematologic malignancy in our cohort, the median mutation burden is <0.1 mutations per megabyte, which is less than that in reported AML and MDS cohorts. Meanwhile, 9 samples from 8 patients with myeloid malignancies in our cohort showed mutation burden close to that in the TCGA AML cohort (supplemental Figure 5A).
Between the top bar graph and the middle heatmap in Figure 3A are heatmaps for patient age, sex, mean platelet volume, the International Society on Thrombosis and Haemostasis Bleeding Assessment Tool (a tool to record both the presence and the severity of bleeding symptoms in patients) score,30 platelet count, somatic mutation data type (whether fibroblast ES data are available), and RUNX1 mutation type. The existence of any correlations between these measures and the detected somatic mutations is further depicted in supplemental Figure 4, by sorting the landscape heatmap with different annotation items. Total somatic mutation number and CHIP gene mutations most frequently seen in the general population (TET2 and DNMT3A) and in high-risk genes (such as KRAS, NRAS, PHF6, ZRSR2, and SF3B1), all trend up with increasing age (supplemental Figure 4B). BCOR mutations were significantly enriched in patients aged between 20 and 60 years (supplemental Figure 4B) and correlated with lower platelet count (supplemental Figure 4D) and mean platelet volume level (supplemental Figure 4E). Patients with BCOR mutations tended to have low platelet count (supplemental Figure 4D). With current data, we did not find correlations between somatically mutated genes and RUNX1 mutation types or sex.
The bottom bar plot of Figure 3A shows the types of base substitutions associated with the somatic mutations. C>T and T>C transitions are more common than transversions.
Pathway and gene ontology analyses of somatically mutated genes showed enrichment of regulation of histone methylation (also seen in CHIP studies10,15). Highly related pathways also include RAS, PI3K-AKT, MAPK, and interleukin-6 (IL-6) signaling, which are related to inflammation (Figure 3C). In addition, mutations were enriched in genes with hemostasis functions and genes transcriptionally regulated by RUNX1.
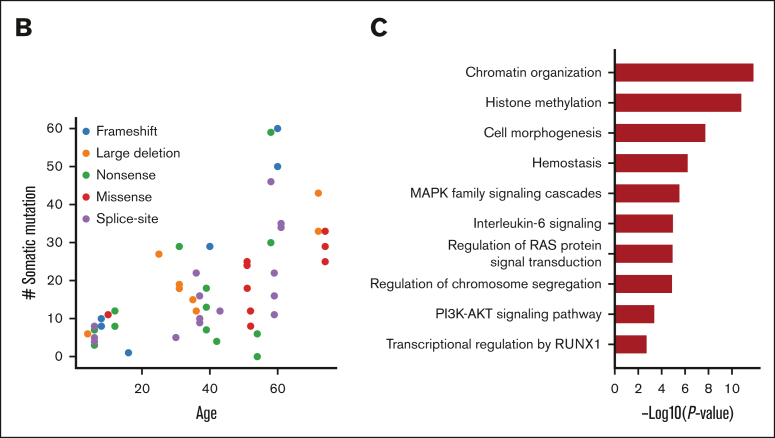
Recurrent somatic mutations in NFE2
As mentioned earlier, we found 7 recurrently mutated genes in the somatic mutation landscape besides the CL genes. Notably, NFE2 was mutated in 3 unrelated patients, including 2 nonsense mutations and 1 missense mutation in the important basic region leucin zipper domain (Figure 4A). NFE2 somatic mutations have been reported in an FPDMM case report31; it encodes a transcription factor involved in megakaryocyte development and platelet production,32,33 which is mutated in 0.6% to 1.3% of patients with MDS, 0.5% of those with leukemia, and 1.6% of pediatric patients with AML34,35 (Figure 4B). However, NFE2 has not been reported as a gene associated with CH. Reported functional studies also indicate that NFE2 is a downstream target of RUNX1.36 Published chromatin immunoprecipitation sequencing data37, 38, 39 showed a strong RUNX1 binding signal in the promoter region of NFE2 in CD34+ hematopoietic stem and progenitor cells and umbilical cord blood–derived megakaryocytes (Figure 4C). In our cohort, 2 of the NFE2 mutation carriers had already developed premonoclonal gammopathy of undetermined significance and myeloma, respectively. Our data suggest that somatic NFE2 mutations might be related to FPDMM disease progression.
Figure 4.
NFE2 somatic mutations in FPDMM and hematologic malignancies. (A) Lollipop plot of NFE2 somatic mutations in NIH FPDMM cohort. Based on ACMG criteria, both nonsense mutations are LP (p.Leu57ArgfsTer27: PVS1 and PM2; p.Gln324Ter: PVS1 and PM2). The missense mutation, p.Asn276His, is considered as a VUS (PP3, PM2, and BP1) based on the PhyloP100way score but pathogenic according to in silico prediction (P/LP: 21/27; uncertain: 6/27). (B) Somatic alteration frequency of NFE2 gene in AACR GENIE data set and cBioportal myeloid neoplasm data set. (C) ChIP-seq data on RUNX1 binding in NFE2 locus in CD34+ cells from 4 publicly available data sets.37, 38, 39 Binding profile data were downloaded from CODEX database.40 AACR GENIE, American Association for Cancer Research Genomics Evidence Neoplasia Information Exchange; HSPCs, hematopoietic stem and progenitor cells; UCB, umbilical cord blood.
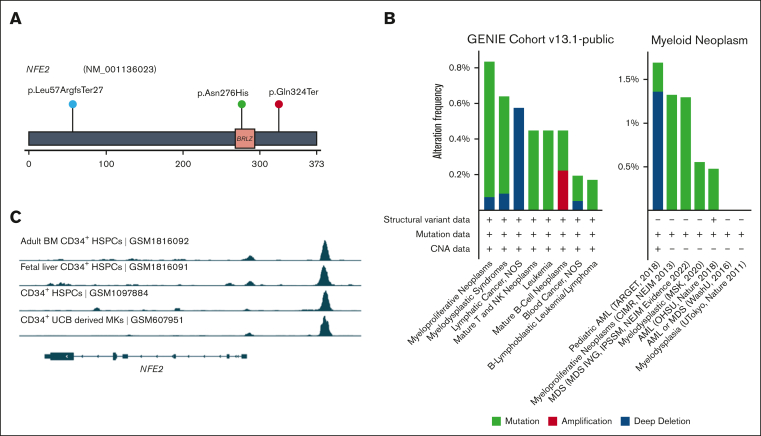
Increased frequency of CH in patients with FPDMM
It has been reported that patients with FPDMM could develop detectable CH with a cumulative risk of >80% by 50 years of age,16 which is far younger than the population average,41 and that detection of these clones may help inform risk of developing hematologic malignancy.42 We set out to determine the frequency of CH in our cohort by comparing our data with those of the population cohort reported by the Trans-Omics for Precision Medicine (TOPMed) research program.27 To meet the criteria applied to the TOPMed study, only somatic mutations detected in the previously reported 74 CHIP genes27 with VAF>5% were included for the comparison (Figure 5A; supplemental Figure 6). Fourteen of 51 (27.5%) patients without hematologic malignancy in the FPDMM cohort have mutations in 11 CHIP genes at VAF > 5%. This frequency is significantly higher (2-tailed z-score test, z = 8.138; P < .00001) than that of the general population (4.3%).27 Moreover, 13 of the 14 (92.9%) patients without any hematologic malignancy with CHIP gene mutations were aged <65 years, with a median age of 42 years, and the youngest patient was aged only 13 years. In the general population, only 10% of people aged >65 years and 1% <50 years were reported to carry CHIP gene mutations. In our cohort, CHIP gene mutations were detected in 1 of 3 patients aged >65 years, 9 of 46 patients (19.6%) <50 years, and 1 of 20 (5%) <20 years.
Figure 5.
CHIP gene mutations in the NIH FPDMM cohort. (A) Bar graph showing the numbers of somatic mutations detected in 74 CHIP genes listed in the TOPMed study.27 Only those with VAF>5% are included. (B) Mutation signatures43 in patients with or without somatic mutations in the CL gene list. (C) Clinical phenotypes with significant difference between patient groups with or without somatic mutations in the CL gene list.
We determined whether there are differences in mutation signatures43 between patients with CL gene mutations and those without the mutations (Figure 5B; supplemental Figure 5B-C). In both CL+ and CL− groups, at least half of the mutations belonged to single-base substitution (SBS) signatures SBS1 and SBS5, which are both “clock-like” mutations44 that accumulate with time. Interestingly, in the CL+ group, 35% of the mutations were assigned to SBS6, which is associated with defective DNA-mismatch repair44,45; in CL⁻ group, 21.4% of the mutations were related to this but classified as SBS15, a signature that also belongs to the defective DNA-mismatch repair category.
We also compared phenotype data between CL+ and CL− groups (Figure 5C). Patients in the CL+ group had significantly lower platelet counts (P = .007) and higher the International Society on Thrombosis and Haemostasis Bleeding Assessment Tool scores (P = .045) than patients in the CL− group. Patients in the CL+ group had lower numbers of CD34+ cells in the BM (P = .012), but a higher proportion of these cells also expressed CD123, which is overexpressed in many hematologic malignancies, including 80% of AML and B-cell ALL.46 Moreover, patients in the CL+ group had significantly higher blood immunoglobulin A level (P < .001), which may lead to increased proinflammatory cytokine production through the activation of Fc fragment of IgA receptor-expressing immune cells.47,48 Interestingly, patients in the CL+ group were significantly older than those in the CL− group (average age of 40 years and 22 years, respectively; P < .01). On the other hand, both groups had balanced male-to-female ratio, and no significant difference was observed in RUNX1 mutation types.
Dynamic changes of somatic mutations over time
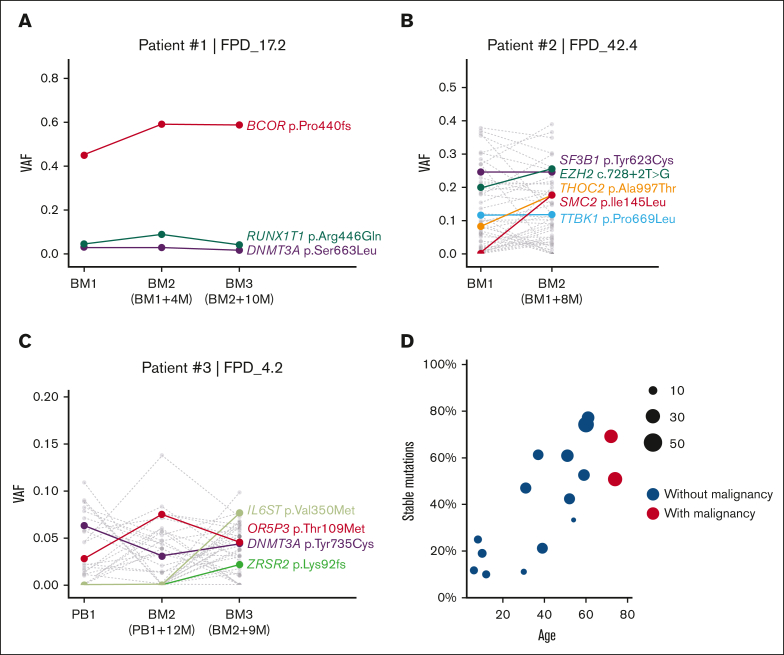
Multiple patients in our cohort had completed their second or third annual visits, and we sequenced their samples to monitor the dynamic changes in their somatic mutations. We observed a patient (patient 1) with a stable dominant clone (clones detected in ≥2 consecutive time points with relatively stable VAFs) characterized by a high VAF BCOR mutation (Figure 6A). Additional mutations in DNMT3A and RUNX1T1 also remained stable at low VAF. Patient 2, who already developed MDS with ring sideroblasts, had a splicing factor SF3B1 mutation, which was stable at 25% VAF (Figure 6B). Mutations in SF3B1 were reported in multiple patients with chronic lymphocytic leukemia and MDS.49 However, VAFs of THOC2 and SMC2 mutations increased significantly at the second visit for patient 2 (Figure 6B). THOC2 is a member of the transcription-export complex, which is indispensable for messenger RNA export50; SMC2 is vital for the structural maintenance of chromosomes.51 The combined annotation–dependent depletion Phred-score of THOC2 and SMC2 mutations are 32 and 29.2, respectively, both predicting a highly deleterious risk. Similarly, we observed rising clones with potential risk in patient 3 (Figure 6C). In the third yearly BM sample, we detected a new frameshift mutation in ZRSR2, which may cause dysregulated RNA splicing,52 and a new somatic mutation in IL6ST, which encodes a signal transducer shared by multiple cytokines, including IL-6 and leukemia inhibitory factor.53 Sequential data on more patients are shown in supplemental Figure 7.
Figure 6.
Somatic mutation VAF changes over time and age. (A-C) Somatic mutation VAF changes across samples collected at different time points. Colored dots and lines show mutations in CL and leukemia genes; gray dots and lines represent other somatic mutations. (D) Correlation between patient’s age and percentage of stable somatic mutations (present in at least 2 time points with VAF > 0.01) in all somatic mutations. Dot size indicates the total somatic mutation number in this patient. BM, bone marrow; PB, peripheral blood.
Although comparing multi-timepoint mutations in our cohort, we observed a pattern that younger patients usually have fewer stable clones over time (fewer somatic mutations that could be detected in multiple time point samples). As shown in Figure 6D and supplemental Figure 8A, fewer mutations were stable (present in at least 2 time points at VAF > 0.01) in patients aged <30 years. In contrast, the fraction of stable mutations increased with age, suggesting the presence of more stable clones in older patients.
Additional genomic risk factors in FPDMM
Besides somatic mutations identified by ES, we also investigated other genomic alterations as potential risk factors cooperating with RUNX1 mutations for malignant transformation.
With SNP-array–based CNV analysis, we detected increased frequency of CNVs in patients with FPDMM. Specifically, we identified CNVs in 10 of 25 (40%) patients without hematologic malignancies (supplemental Table 4), whereas we detected a CNV in only 1 of 8 family controls (12.5%). For most patients with CNVs, there were no more than 2 CNVs, and only limited genomic regions were affected. By cytogenetics analysis, only 1 of the 51 analyzed patients without a hematologic malignancy had an abnormal karyotype (a marker chromosome in the BM cells; supplemental Table 4). In addition, fusion-gene analysis did not reveal any in-frame fusion events in the 21 patients for whom we had RNA-seq data.
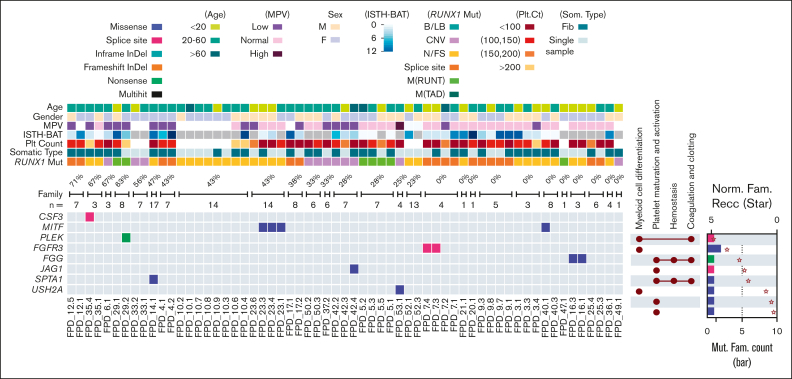
We have also cataloged and analyzed germline variants in the enrolled families in an attempt to identify modifiers of disease. We focused on genes related to myeloid cell differentiation and hemostasis functions. In 9 families, we detected germline variant either classified as P/LP by Clinvar or classified as P/LP following ACMG criteria. Affected genes included CSF3, MITF, PLEK, FGFR3, FGG, JAG1, SPTA1, and USH2A (Figure 7). We also included the percentage of patients in each family who developed hematologic malignancies to highlight genes enriched in these high penetrance families.
Figure 7.
Heatmap of P/LP germline variants in genes related to myeloid cell development and hemostasis. Demographic and clinical data are depicted (top), including percentages of affected family members who developed hematologic malignancy in each family (“n=” denotes the total numbers of family members). The UpSet plots on the right of the heat map show the related functional annotations for each gene. The dots indicate that the gene in that row is involved in the functional categories listed at the top. The bar plots on the far-right show numbers of families (Mut. Fam. Count) that carry germline mutations in each gene (scale labeled at the bottom of the plot). Protein size–normalized family number (Norm. Fam. Recc.) are shown with star sign, to provide the affected frequency in a normalized level (scale labeled at the top of the plot). The color codes for mutation types (both the heat map and the bar plot) and for the clinical and demographic data are listed below the heat map. B/LB, benign/likely benign; CNV, copy number variation; F, remale; Fib, Fibroblast; M(RUNT), mutation in RUNT domain; M, male; M(TAD), mutation in transactivation domain; N/FS, nonsense/frame shift.
Because disease modifiers are not essentially P/LP mutations, we also demonstrated the distribution of variants predicted to be deleterious by in silico modeling, although these were not necessarily considered pathogenic based on the ACMG criteria with respect for monogenic disease17 (supplemental Figure 9A). For example, we found TNFSF9 variants in 3 families; the protein encoded by this gene belongs to the tumor necrosis factor ligand family, and it correlated with platelet phenotypes in genome-wide association studies.54 We found germline variants in genes closely related to myeloid malignancies, such as GATA2, FGFR3, and ITGB2, and in genes involved in biological functions related to the phenotype of FPDMM, such as SERPINA10, SRF, and VWF.
In our cohort, we identified predicted-deleterious germline variants in Fanconi anemia genes. Among the 7 families with these variants, 4 showed a high rate of hematologic malignancies within their family (supplemental Figure 9B; supplemental Table 5). Genes in the RTK-RAS-PI3K pathway also showed germline alterations (supplemental Figure 9C; supplemental Table 5). Our current cohort size is still limited to the power of discovering the influence of germline variants, and the exact significance of these germline variants is unknown; however, it is possible that stronger associations will be detected in the future with more participants enrolled through our longitudinal study.
Discussion
In our cohort, germline variants were found to disrupt RUNX1 mainly in following 2 ways: (1) loss-of-function variants truncating or completely deleting the RUNX1 protein and (2) missense variants affecting critical functional domains. In addition, we have observed splice donor and acceptor sites as mutation hotspots in our patients, which lead to the usage of cryptic splice sites and alternative splicing events, resulting insertions, deletions, frameshifts, and truncations. Because of the deleterious nature of the pathogenic germline RUNX1 variants and the presence of mutational hot spot, therapeutic strategies can be envisioned to increase or restore RUNX1 expression from the normal allele and block or repair hot spot mutations through antisense oligo, CRISPR, or other targeted strategies.
Our somatic mutation landscape demonstrated that approximately half of the patients with FPDMM have at least 1 somatic mutation in a CL gene. Compared with patients without these mutations, the affected group showed significantly different phenotypes, including lower platelet count, lower percentage of CD34+ cells in the marrow, and higher blood immunoglobulin A level. These findings are neither related to the difference in the sex ratio nor to different RUNX1 mutation types, but the CL+ group is associated with more advanced age. Functional enrichment analysis of the mutated genes identified several essential functions or pathways related to inflammation and RAS/PI3K-AKT-mTOR/MAPK. Mutation signatures also indicate that the affected group has a higher signal linked to DNA-mismatch repair. Besides CHIP genes frequently seen in the general population, BCOR is the top mutated gene in patients with FPDMM.27 Moreover, BM samples from 4 patients without hematologic malignancy harbored >1 BCOR mutation. In 1 of them, there were 5 different BCOR mutations, with VAFs ranging from 2.3% to 40%. From our data, most patients with BCOR mutations have not developed hematopoietic malignancy, even for those with BCOR mutations at relatively high VAFs. Therefore, the mutation mechanism and the impact of BCOR mutations on malignant transformation need further investigation. In addition, NFE2, a RUNX1 target gene that encodes a regulator of megakaryocyte differentiation, was found to be frequently mutated. Overall, patients with FPDMM seemed to have a distinct somatic mutation landscape, which might have been shaped by RUNX1 haploinsufficiency caused by the germline mutations, as recently described for Shwachman-Diamond Syndrome.55
Our study identified somatic mutations in several genes that were already reported in MDS or AML, such as NRAS, KRAS, SF3B1, PHF6, and ZRSR2. Interestingly, we found somatic NFE2 mutations in 3 unrelated patients: 1 of them developed a lymphoid malignancy. As a gene downstream of RUNX1 and involved in megakaryocyte development, NFE2 has not received much attention in the context of FPDMM, except for a single recent case report.31 It could be an important player and, therefore, a candidate to study its role in FPDMM in the context of hematologic malignancy development.
We do not have enough evidence to link other recurrently mutated genes with FPDMM. But for example, histone demethylase KDM3A is an essential member of the JAK2-STAT3 signaling pathway56 that might be related to hematologic malignancy. We may find more recurrently mutated genes when we gather data of more patients. Therefore, we will pay attention to these genes and further confirm their possible link to the disease development with additional data and through in vitro and in vivo functional studies. LRP1B is a large (90 exons) gene, so the detected LRP1B mutations may not be significant. However, we did not detect mutations in several other large genes, such as BRCA1, APC, and NF1.
In previous studies, somatic mutations in RUNX1 were found to be common in patients with FPDMM who have developed malignancies.4 However, for the 12 patients with hematologic malignancy in our cohort, only 1 carried a RUNX1 somatic mutation, a large deletion. This frequency is far below the reported frequency (>40%). It is unclear why we have not seen more somatic mutations in RUNX1 in our cohort. It would be interesting to see whether we detect more RUNX1 somatic mutations as we expand our cohort and continue our longitudinal study of the patients.
Longitudinal tracking of somatic mutations can help us monitor dynamic changes of clonal expansion and transformation to malignancies. We observed multiple patients with 2 or 3 time points; some of them had somatic mutations with stable VAFs, whereas others have mutations with fluctuating VAFs. Interestingly, younger patients tended to have higher VAF fluctuations between time points, whereas older patients tended to have more stable mutations. The clinical significance of these findings is unclear and, hopefully, will become more apparent with longer follow-up of these patients.
In addition to somatic mutations, deleterious germline variants in genes related to hematologic malignancies may also increase the risk of malignancy development in patients with FPDMM. However, determining which candidate germline variants are relevant to pathogenesis and progression of FPDMM is difficult without experimental validation or statistical power. We believe that after accumulating more data from the patients and their families and performing experiments for functional confirmation, we will be able to identify germline modifiers that may stratify the risk for patients with FPDMM.
Early and accurate detection of disease progression in FPDMM is imperative for clinical management and improving outcomes. We will continue following enrolled patients to collect more “snapshots” of their genomes and clinical phenotypes, with the goal of identifying risk factors as early as possible. Hopefully, our study will determine the significance of somatic mutations for malignant transformation, which may lead to discoveries of biomarkers for disease progression in FPDMM, eventually benefiting clinical management for patients with FPDMM.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors are grateful to patients and their family members for their participation in this clinical study.
This work was supported by the Intramural Research Programs of the National Human Genome Research Institute (NHGRI) and National Cancer Institute, National Institutes of Health (NIH). The authors thank the NIH Intramural Sequencing Center, NHGRI Genomics, Microarray, Bioinformatics, Cytogenetics Core, and National Institute of Arthritis and Musculoskeletal and Skin Diseases Genomic Technology Section for generating genomic data. The authors thank NIHCC for conducting clinical procedures, pathology evaluations, and laboratory tests. The authors also thank the RUNX1 Research Program, a patient advocacy nonprofit, for their help with recruiting study subjects and for travel support for international patients. This work used the computational resources of the NIH HPC Biowulf cluster.
Authorship
Contribution: K.Y. and P.P.L. designed the analysis strategy and wrote the manuscript; K.Y., M.M., E.B., and J. Diemer performed the experiments; K.Y. and R.S. analyzed the data; N.D. provided genetic counseling; J. Davis, N.D., M.M., and L.C. enrolled patients; M.M. and L.C. provided clinical care; L.C. served as a medical director; P.P.L. served as a protocol principal investigator; and all authors discussed and revised the manuscript.
Footnotes
All data described in this manuscript are based on reference hg38 coordinates.
The raw ES, RNA-seq and SNP-array data have been deposited in the database of Genotypes and Phenotypes (dbGaP) (accession number phs003075).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.de Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129(15):2061–2069. doi: 10.1182/blood-2016-12-689109. [DOI] [PubMed] [Google Scholar]
- 2.Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 4.Brown AL, Arts P, Carmichael CL, et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv. 2020;4(6):1131–1144. doi: 10.1182/bloodadvances.2019000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godley LA. Inherited predisposition to acute myeloid leukemia. Semin Hematol. 2014;51(4):306–321. doi: 10.1053/j.seminhematol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Brown AL, Churpek JE, Malcovati L, Döhner H, Godley LA. Recognition of familial myeloid neoplasia in adults. Semin Hematol. 2017;54(2):60–68. doi: 10.1053/j.seminhematol.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Simon L, Spinella JF, Yao CY, et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood. 2020;135(21):1882–1886. doi: 10.1182/blood.2019003357. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Yang W, Devidas M, et al. Germline RUNX1 variation and predisposition to childhood acute lymphoblastic leukemia. J Clin Invest. 2021;131(17) doi: 10.1172/JCI147898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antony-Debré I, Duployez N, Bucci M, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016;30(4):999–1002. doi: 10.1038/leu.2015.236. [DOI] [PubMed] [Google Scholar]
- 10.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465) doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157–170. doi: 10.1016/j.stem.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaiswal S, Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17(3):137–144. doi: 10.1038/s41569-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaiswal S. Clonal hematopoiesis and nonhematologic disorders. Blood. 2020;136(14):1606–1614. doi: 10.1182/blood.2019000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Churpek JE, Pyrtel K, Kanchi KL, et al. Genomic analysis of germ line and somatic variants in familial myelodysplasia/acute myeloid leukemia. Blood. 2015;126(22):2484–2490. doi: 10.1182/blood-2015-04-641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham L, Merguerian MD, Calvo KR, et al. Natural history study of patients with familial platelet disorder with myeloid malignancy. Blood. Published online 22 September 2023 doi: 10.1182/blood.2023019746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales J, Pujar S, Loveland JE, et al. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022;604(7905):310–315. doi: 10.1038/s41586-022-04558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oetjen KA, Lindblad KE, Goswami M, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight. 2018;3(23) doi: 10.1172/jci.insight.124928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4) doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaud J, Wu F, Osato M, et al. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99(4):1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 24.De Rocco D, Melazzini F, Marconi C, et al. Mutations of RUNX1 in families with inherited thrombocytopenia. Am J Hematol. 2017;92(6):E86–E88. doi: 10.1002/ajh.24703. [DOI] [PubMed] [Google Scholar]
- 25.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10(11):1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bick AG, Weinstock JS, Nandakumar SK, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586(7831):763–768. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network. Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adler M, Kaufmann J, Alberio L, Nagler M. Diagnostic utility of the ISTH bleeding assessment tool in patients with suspected platelet function disorders. J Thromb Haemost. 2019;17(7):1104–1112. doi: 10.1111/jth.14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duarte BKL, Yamaguti-Hayakawa GG, Medina SS, et al. Longitudinal sequencing of RUNX1 familial platelet disorder: new insights into genetic mechanisms of transformation to myeloid malignancies. Br J Haematol. 2019;186(5):724–734. doi: 10.1111/bjh.15990. [DOI] [PubMed] [Google Scholar]
- 32.Marcault C, Zhao LP, Maslah N, et al. Impact of NFE2 mutations on AML transformation and overall survival in patients with myeloproliferative neoplasms. Blood. 2021;138(21):2142–2148. doi: 10.1182/blood.2020010402. [DOI] [PubMed] [Google Scholar]
- 33.Jutzi J S, Basu T, Pellmann M, et al. Altered NFE2 activity predisposes to leukemic transformation and myelosarcoma with AML-specific aberrations. Blood. 2019;133(16):1766–1777. doi: 10.1182/blood-2018-09-875047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.AACR Project GENIE Consortium AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Schwemmers S, Hexner EO, Pahl HL. AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood. 2010;116(2):254–266. doi: 10.1182/blood-2009-11-254664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J, Liu X, Li D, et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell. 2016;36(1):9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beck D, Thoms JAI, Perera D, et al. Genome-wide analysis of transcriptional regulators in human HSPCs reveals a densely interconnected network of coding and noncoding genes. Blood. 2013;122(14):e12–22. doi: 10.1182/blood-2013-03-490425. [DOI] [PubMed] [Google Scholar]
- 39.Tijssen MR, Cvejic A, Joshi A, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20(5):597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sánchez-Castillo M, Ruau D, Wilkinson AC, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2015;43:D1117–D1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergstrom EN, Huang MN, Mahto U, et al. SigProfilerMatrixGenerator: a tool for visualizing and exploring patterns of small mutational events. BMC Genomics. 2019;20(1):685. doi: 10.1186/s12864-019-6041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meier B, Volkova NV, Hong Y, et al. Mutational signatures of DNA mismatch repair deficiency in C. elegans and human cancers. Genome Res. 2018;28(5):666–675. doi: 10.1101/gr.226845.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Testa U, Pelosi E, Frankel A. CD 123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomark Res. 2014;2(1):4. doi: 10.1186/2050-7771-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen IS, Baeten DLP, den Dunnen J. The inflammatory function of human IgA. Cell Mol Life Sci. 2019;76(6):1041–1055. doi: 10.1007/s00018-018-2976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen IS, Hoepel W, Zaat SAJ, Baeten DLP, den Dunnen J. Serum IgA immune complexes promote proinflammatory cytokine production by human macrophages, monocytes, and Kupffer cells through FcalphaRI-TLR cross-talk. J Immunol. 2017;199(12):4124–4131. doi: 10.4049/jimmunol.1700883. [DOI] [PubMed] [Google Scholar]
- 49.Foy A, McMullin MF. Somatic SF3B1 mutations in myelodysplastic syndrome with ring sideroblasts and chronic lymphocytic leukaemia. J Clin Pathol. 2019;72(11):778–782. doi: 10.1136/jclinpath-2019-205895. [DOI] [PubMed] [Google Scholar]
- 50.Viphakone N, Hautbergue GM, Walsh M, et al. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7(5):311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 52.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 53.Murakami M, Kamimura D, Hirano T. Pleiotropy and specificity: insights from the interleukin 6 family of cytokines. Immunity. 2019;50(4):812–831. doi: 10.1016/j.immuni.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 54.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kennedy AL, Myers KC, Bowman J, et al. Distinct genetic pathways define pre-malignant versus compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. Nat Commun. 2021;12(1):1334. doi: 10.1038/s41467-021-21588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H, Kim D, Choi SA, et al. KDM3A histone demethylase functions as an essential factor for activation of JAK2-STAT3 signaling pathway. Proc Natl Acad Sci U S A. 2018;115(46):11766–11771. doi: 10.1073/pnas.1805662115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.