Abstract
Cyclin-dependent kinases (CDKs) drive the cell cycle, central to which is the accurate control of chromosome replication. In Saccharomyces cerevisiae, six closely related B-type cyclins (Clb1–6) drive the events of S phase and mitosis. Either Clb5 or Clb6 can activate early-firing replication origins, whereas only Clb5 can activate late origins. Clb1–4 are expressed later in the cell cycle. Whether Clb cyclins differ only in timing of expression, or else impart different kinase specificities is under ongoing investigation. This study shows that the expression of Clb2 during S phase in cells lacking Clb5 failed to rescue late origin activation. Early expression of Clb2 in cells lacking both Clb5 and Clb6 did not activate early origins on schedule to restore the correct S phase entry time. Therefore, Clb2 cannot drive timely activation of either early or late replication origins, demonstrating that Clb2-directed CDK has a specificity distinct from that driven by Clb5 and Clb6.
INTRODUCTION
Nine cyclins are expressed in an ordered sequence and associate with the cyclin-dependent kinase (CDK) Cdc28 to drive the yeast cell cycle. The three G1 (Cln) cyclins initially switch on a series of six B-type (Clb) cyclins, beginning with the expression of Clb5 and Clb6 during S phase, followed by Clb3 and Clb4 in early G2, and finally Clb1 and Clb2 later in G2 (reviewed in Nasmyth, 1996). Clb5 and Clb6 drive chromosome replication (Figure 1A) (Epstein and Cross, 1992; Kühne and Linder, 1993; Schwob and Nasmyth, 1993). Either Clb5 or Clb6 is able to activate early-firing replication origins but only Clb5 can activate late replication origins, so that S phase in clb5 mutant cells is prolonged due to the failure of late origin activation (Figure 1B) (Donaldson et al., 1998). Clb1,2,3 and 4 normally promote mitotic events. However, in the absence of Clb5 and Clb6, Clb1,2,3 and 4 can jointly take over the role of activating S phase by firing both early and late origins (Figure 1C). Such redundancy has led to questions concerning the extent to which the various B-type cyclin proteins are functionally interchangeable. This study addresses whether cells that express the mitotic cyclin Clb2 in place of one or both S phase cyclins can fire their replication origins correctly.
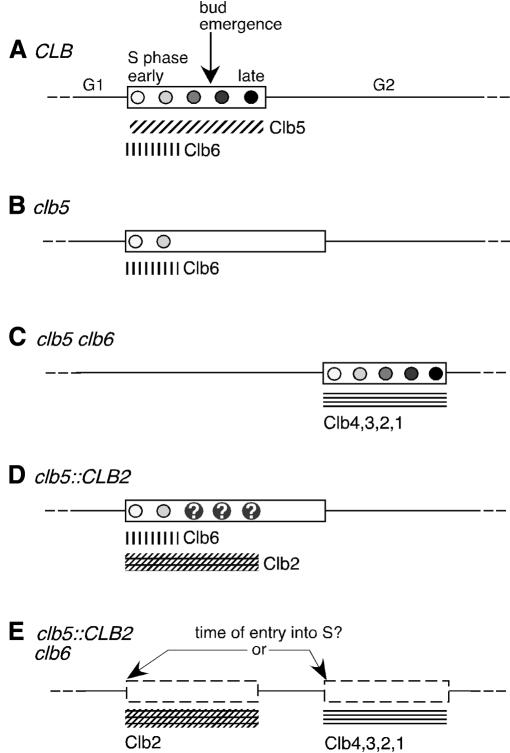
Fig. 1. Summary of known roles of B-type cyclins in origin activation and outline of the experiments described. (A–E) show time lines representing the cell cycle in various strains. S phase is shown as a rectangle; shaded circles represent replication origin activation events, with light circles as early-firing origins and darker circles as later-firing origins. Hatched bars refer to the function of various cyclins in activating replication origins. (A) Wild-type (CLB) cell cycle. Clb5 can activate both early and late origins whereas Clb6 is only capable of activating early origins. (B) In clb5 cells late origins fail to fire normally and late replicons are instead replicated ‘passively’ by forks from early origins, with the result that S phase is prolonged. (C) clb5 clb6 cells are delayed in entering S phase, because replication can only begin once Clb1–4 activity has accumulated. (D) This study addresses whether late origins fire (question marks) in a strain that expresses Clb2 in place of and at the normal time of Clb5 (the clb5::CLB2 strain). (E) In the second part of this study, I investigate whether early expression of Clb2 in the absence of Clb5 and Clb6 fires early origins to allow cells to enter S phase on time.
RESULTS AND DISCUSSION
Early expression of Clb2 cannot activate late origins
Cross et al. (1999) recently constructed a strain in which the CLB5 open reading frame (ORF) is precisely replaced with that of CLB2. The resulting clb5::CLB2 construct expresses CLB2 message under the control of the CLB5 promoter, causing artificially early accumulation of Clb2–Cdc28 kinase activity instead of, and at the normal time of Clb5–Cdc28 activity. The ability of Clb2–Cdc28 activity to fire late replication origins can therefore be tested by analysing late origin firing in the clb5::CLB2 strain (Figure 1D).
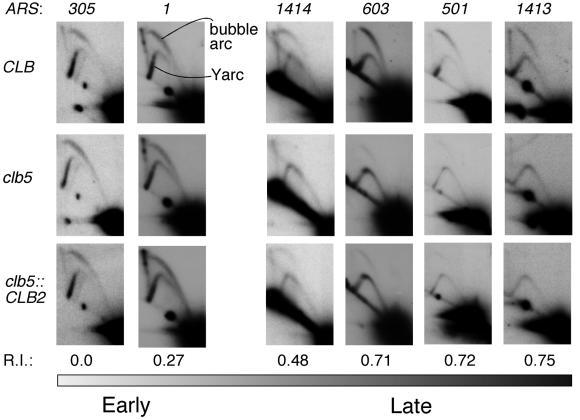
Late origin firing was examined in the clb5::CLB2 strain (Figure 2) using two-dimensional agarose gel electrophoresis (Brewer and Fangman, 1987; Friedman and Brewer, 1995). This technique separates replication intermediates so that DNA fragments containing bubble structures (and therefore active replication origins) form a distinctive ‘bubble arc’ (see ARS1 panel in Figure 2). Replication of the fragment by forks originated elsewhere instead results in a ‘Y arc’.
Fig. 2. 2D gel analysis of early and late replication origin use. Origin use was examined in CLB, clb5 and clb5::CLB2 strains (top, middle and bottom panels, respectively). The presence of a bubble arc indicates that the origin is activated, while the Y arc represents the proportion of fragments replicated passively. Early origin use appears normal in all three strains. Late origin firing is greatly diminished in clb5 strains. Late origin firing in clb5::CLB2 strains appears defective as in the clb5 mutant. Names of the origins are given above the panels. R.I. value is a measure of origin activation time, with 0.0 representing a very early origin and 0.75 one of the latest known origins (Friedman et al., 1996).
Early replication origin firing appeared normal in CLB (wild type), clb5 and clb5::CLB2 strains, as illustrated in Figure 2 for the early origins ARS305 and ARS1. ARS603 best exemplifies the results obtained for late origins. Clear bubble arcs confirmed late origin activity in the CLB strain, but as described previously (Donaldson et al., 1998), late origin firing was greatly diminished in the clb5 strain. Bubble arcs were also very faint in the clb5::CLB2 strain, indicating that, in general, late origins were not activated by premature Clb2 activity replacing that of Clb5. Expression of Clb2 during S phase, therefore, did not rescue the late origin-firing defect of the clb5 mutant.
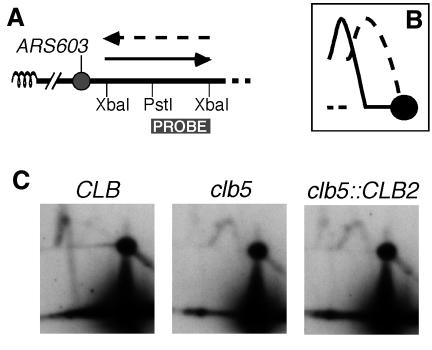
To allow quantitative estimation of the activation of a late origin in the clb5::CLB2 strain, the direction of replication fork movement was analysed at a locus to the right of the late replication origin ARS603 using the modification to the 2D-gel procedure described previously (Friedman and Brewer, 1995; Friedman et al., 1997). The chromosomal location of ARS603 makes it convenient for this analysis; it is the leftmost efficient origin on chromosome VI so that replication forks progressing rightward through sequences to the right of ARS603 must have been initiated at that origin (Figure 3A). If ARS603 is not activated this chromosome region is instead replicated ‘passively’ by a leftward-moving replication fork initiated at a more centromere-proximal (presumably early) origin (Figure 3A). The cartoon in Figure 3B illustrates the 2D gel arcs expected if ARS603 is active or inactive. Results of the analysis in CLB, clb5 and clb5::CLB2 strains are shown in Figure 3C. As in the clb5 strain, in the clb5::CLB2 strain the predominant arc corresponded to a leftward-moving fork. ARS603 was therefore inactive and the region was instead replicated passively by a leftward-moving replication fork in the majority of clb5::CLB2 cells.
Fig. 3. Analysis of replication fork direction close to the late origin ARS603. (A) Chromosome configuration of the left arm of chromosome VI. If ARS603 is activated, the XbaI fragment immediately to its right is replicated by the rightward-moving fork from ARS603 (solid arrow). ARS603 is the closest efficient origin to the left telomere (coiled line), so that if ARS603 is not activated the XbaI fragment is instead replicated by a leftward-moving fork (dashed arrow). The direction of replication fork movement was analysed as described by Friedman and Brewer (1995), using PstI for the in-gel digestion and probing for the larger PstI–XbaI fragment. (B) The solid curve illustrates the Y arc expected if replication forks move rightward through the XbaI fragment (i.e. ARS603 active). The dashed curve shows the expected Y arc if forks move leftward (i.e. ARS603 inactive). (C) Fork-direction analysis within the XbaI fragment in CLB, clb5 and clb5::CLB2 strains. Quantitation of these blots (as described by Friedman et al., 1997) allowed estimation that ARS603 fires in 74% of CLB cell cycles, 23% of clb5 cell cycles and 32% of clb5::CLB2 cell cycles. The value obtained for CLB may be an underestimate of actual origin firing, because the minor arc was barely visible above background. Similarly, the value for clb5 may represent an overestimation. Nevertheless, the values are generally consistent with those found previously (Friedman et al., 1997; Donaldson et al., 1998).
The arc corresponding to the rightward replication fork from ARS603 did, however, appear slightly increased in the clb5::CLB2 when compared with the clb5 strain. To estimate origin firing of ARS603 in the three strains, the proportion of forks moving in either direction was quantitated as described in Friedman and Brewer (1995) (see also Figure 3 legend). The results indicated that ARS603 origin firing was slightly restored in the clb5::CLB2 cells when compared with clb5 (firing in ∼23% of clb5 and ∼32% of clb5::CLB2 cells). Results from the fork-direction gel analysis of ARS603 firing in the clb5::CLB2 strain, therefore, confirmed those from the conventional 2D gels.
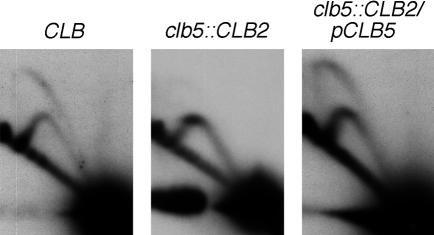
Overexpression of Clb2 has been found to reduce the message levels of a number of S phase genes including RNR1, which encodes ribonucleotide reductase (Spellman et al., 1998). To investigate whether this effect might cause the failure of late origin activation in the clb5::CLB2 strain, firing of ARS603 was tested in clb5::CLB2/pCLB5 cells, which contain a plasmid-borne copy of wild-type CLB5 under its own promoter. Late origin firing was restored in this strain (Figure 4), ruling out the possibility that the late origin firing defect in the clb5::CLB2 strain was the result of such a mechanism.
Fig. 4. The late origin ARS603 is active in a clb5::CLB2/pCLB5 strain. 2D gel analysis of replication intermediates in asynchronously growing CLB (left), clb5::CLB2 (centre) and clb5::CLB2/pCLB5 (right) strains.
The 2D gel analyses indicated that late origin firing remains deeply defective when Clb2 is expressed in place of Clb5, and that premature expression of Clb2, therefore, cannot fulfil the late origin-firing function of Clb5. Quantitative estimation using fork-direction gel analysis showed that early Clb2 expression results in only slight rescue of the late origin defect of a clb5 mutant. These results are also in close agreement with those described by Cross et al. (1999). The lengthened S phase of clb5 deletion mutants (Figure 1B) is manifest in asynchronous cultures as an accumulation of cells with DNA content intermediate between that of G1 and G2. Cross et al. used flow cytometry analysis to show that asynchronously growing clb5::CLB2 cultures have a similar accumulation of S phase cells, indicating that S phase is also lengthened in the clb5::CLB2 strain. Both the analysis in Cross et al. (1999) and the results presented here show that Clb2 is unable to fulfil the S phase function of Clb5.
Early expression of Clb2 does not activate early origins on time
Next, I tested whether prematurely expressed Clb2 can activate early replication origins. clb5 cells begin S phase at the normal time because Clb6 alone is sufficient to activate early replication origins (Figure 1B). clb5 clb6 mutants are dependent on one or more of cyclins Clb1–4 to promote S phase, and are therefore delayed in entering S phase (Figure 1C) (Donaldson et al., 1998). Whether prematurely expressed Clb2 activates early replication origins can therefore be investigated (Figure 1E) by testing whether a clb5::CLB2 clb6 mutant begins S phase on time (like the clb5 strain) or is instead delayed in S phase entry (like clb5 clb6).
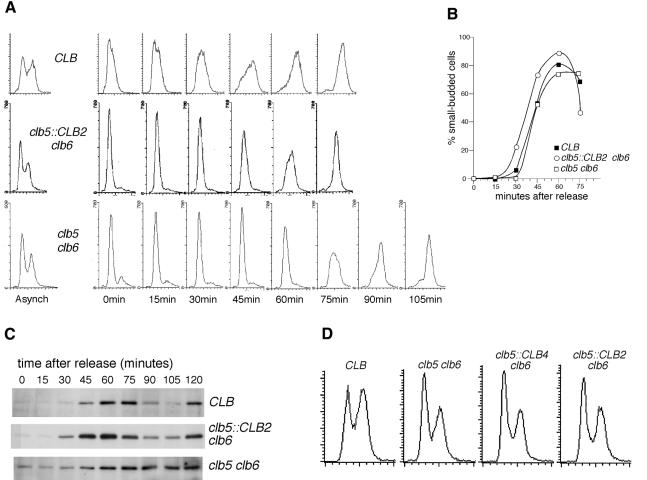
Because of their delayed S phase entry and consequent shorter G2 phase, asynchronously growing cultures of clb5 clb6 cells have a reduced proportion of cells with a G2 DNA content (Figure 5A, left hand panels). clb5::CLB2 clb6 cultures showed a similar depressed G2 peak in their flow cytometry profile, suggesting that they share the delayed S phase entry found in the clb5 clb6 mutant.
Fig. 5. Time of S phase and Clb4 accumulation in CLB, clb5::CLB2 clb6 and clb5 clb6 strains. (A) Flow cytometry analysis of DNA content in asynchronously growing cultures (left panels) and in cultures synchronized by release from α-factor (right panels). Time is minutes after release. (B) Kinetics of bud emergence in (A). (C) Western blot analysis of Clb4-HA accumulation in CLB, clb5::CLB2 clb6 and clb5 clb6 strains, in α-factor release experiments on strains with epitope-tagged Clb4. Bud emergence kinetics were very similar to those in (B). (D) Flow cytometry analysis of asynchronously growing CLB, clb5 clb6, clb5::CLB4 clb6 and clb5::CLB2 clb6 strains.
DNA replication was examined in synchronized cultures (Figure 5A, right hand panels). S phase in the CLB strain began ∼30 min after release from α-factor. The clb5::CLB2 clb6 culture began S phase ∼45 min after release, corresponding to a delay of 15 min when compared with CLB cells. The difference in S phase entry time was not due to faster cell cycle progression of the CLB strain, because budding rates after release were similar for the two strains (Figure 5B). This delay in S phase entry of the clb5::CLB2 clb6 strain showed that early Clb2 expression is inadequate to fire early origins on schedule and restore the normal S phase entry time.
As expected, DNA replication in clb5 clb6 cells was also delayed, by ∼30 min when compared with the CLB strain. The clb5::CLB2 clb6 S phase entry was, therefore, somewhat advanced when compared with clb5 clb6. In other words, early Clb2 expression in a clb5 clb6 background did have some effect on the time of replication initiation. Two possible mechanisms can be envisaged for this advancement of S phase entry time by the clb5::CLB2 construct. Cdc28–Clb2 activity could have some competence for eventual activation of early origins, despite the fact that it is unable to activate them at the correct time. Alternatively, the advanced S phase entry time of clb5::CLB2 clb6 when compared with clb5 clb6 might be an indirect effect of early Clb2 expression—in particular, Clb2 might affect the expression time of other mitotic cyclins that could be involved in activating origins in the absence of S phase cyclins.
To ascertain whether early Clb2 expression affects expression of other cyclins, the kinetics of Clb4 accumulation were compared in CLB, clb5 clb6 and clb5::CLB2 clb6 strains. Western blot analysis of synchronized cultures showed that Clb4 levels are maximal by ∼60 min after release from α-factor in CLB and in clb5 clb6 cells (Figure 5C), whereas in the clb5::CLB2 clb6 strain, Clb4 levels peaked ∼15 min earlier. The data obtained from flow cytometry and western analyses were consistent with the notion that Clb4 (and/or Clb3) might activate S phase in the clb5::CLB2 clb6 and clb5 clb6 strains. To test the competence of Clb4 in activating S phase, I constructed a clb5::CLB4 clb6 strain. This strain lacks both S phase cyclins and has the CLB5 ORF precisely replaced by that of CLB4, so that Clb4 is expressed from the CLB5 promoter. Flow cytometry analysis of asynchronously growing clb5::CLB4 clb6 cells showed a reduced peak of cells with G2 DNA content, very similar to that in the clb5::CLB2 clb6 culture (Figure 5D). This result implied that S phase in the clb5::CLB4 clb6 strain is delayed as in clb5::CLB2 clb6, and S phase entry time is not significantly rescued by early expression of Clb4 protein.
The result obtained from the clb5::CLB4 clb6 strain argues against a substantially better proficiency of Clb4 than Clb2 in promoting timely S phase entry. The advancement of S phase in clb5::CLB2 clb6 cells when compared with a clb5 clb6 strain (Figure 4A) is, therefore, most likely due to a direct effect of early Clb2 expression than to the observed advancement of Clb4 expression in response to early Clb2. These results cannot exclude the possibility that either Clb3- or Clb4-directed Cdc28 does contribute directly to origin firing in clb5::CLB2 clb6 cells—however, a more probable interpretation of the data in Figure 5A is that, despite its incompetence for timely early origin activation, Clb2 can contribute to eventual activation of at least some replication origins in the absence of S phase cyclins.
The experiments described in this study examined the extent to which a mitotic cyclin (Clb2) that is artificially expressed at the normal time of S phase can fire early and late replication origins, and so substitute for the S phase cyclins. The results indicate that premature Clb2 is unable to activate either class of origins at the correct time and efficiency. Late origin firing was almost as defective in the clb5::CLB2 strain as in a clb5 mutant. The marginal improvement in late origin firing efficiency in clb5::CLB2 probably represents a slight competence of Clb2–Cdc28 to fire late replication origins directly. Clb2–Cdc28 also failed to fire early replication origins at the correct time. The clb5::CLB2 construct promotes Clb2–Cdc28 kinase activity at the same time and to similar levels as normal Clb5–Cdc28 activity (Cross et al., 1999). Nevertheless, S phase entry was significantly delayed in the clb5::CLB2 clb6 strain, showing that early Clb2 expression cannot substitute for Clb6 or Clb5 to restore timely early origin firing. Although Clb4 expression was advanced in the clb5::CLB2 clb6 strain, this effect did not appear to cause the advancement in S phase onset when compared with clb5 clb6. The current data instead suggests that Clb2, possibly together with other mitotic cyclins, is unable to fire early origins on time but can eventually activate some replication origins.
In contrast to some previous studies (for example, Amon et al., 1994), the results described here address the capability of cyclins expressed at normal levels to fire origins. Haase and Reed (1999) have succeeded in constructing a strain that is deleted for all six B-type cyclins and maintained by overexpression of CLB1 under galactose promoter control. Time of S phase entry of this clb1,2,3,4,5,6 GAL1-CLB1 strain has not been measured; however, its viability implies that, when overexpressed, the mitotic cyclin Clb1 can drive activation of at least the minimum number of origins required to complete chromosome replication. Overexpression is, however, likely to be essential for Clb1 to fulfil this task, since a clb3,4,5,6 strain is inviable. Clb1 could well share the origin-firing ability shown here for Clb2—that is, defective for timely origin firing but able to promote some eventual chromosome replication.
In general, the presence of multiple B-type cyclins is a complicating factor in experiments aimed at investigating the competence of individual Clb–Cdc28 activities to fire replication origins. Future experiments will test origin activation when single cyclins are expressed in cells lacking all other B-type cyclin activities. While such experiments will be necessary to fully understand cyclin specificity in origin firing, it is already clear from the results presented here that substitution of Clb5 and Clb6 by Clb2 cannot restore a normal S phase.
METHODS
Strain constructions. All strains were derived from BF264-15D (Richardson et al., 1992). Strains 1768-12A and FC12-18 (both CLB), 1768-1A-1 (clb5), 1768-1A-2 (clb5::CLB2) and 1768-6C (clb5 clb6) were gifts from F.R. Cross. Other clb6 derivatives were made from these strains by transformation with a clb6::LEU2 deletion construct as described in Donaldson et al. (1998). pCLB5 (used to make clb5::CLB2/pCLB5) was YCplac111 containing a 3.0 kb BstXI–XbaI CLB5 insert (Schwob and Nasmyth, 1993). The CLB4 gene was HA-tagged at its 3′ terminus by the method of Longtine et al. (1998). A clb5::CLB4 allele was PCR amplified using long primers that included the HindIII site 66 bp upstream of CLB5 or the EcoRI site 22 bp from the CLB5 3′ end. The resulting clb5::CLB4 PCR fragment was HindIII–EcoRI digested and ligated, together with the 812 bp EcoRI–XbaI fragment immediately 3′ to CLB5, into HindIII–XbaI-cut pBluescript KS–. Resulting clones were sequenced and an error-free 2267 bp HindIII clb5::CLB4 fragment was transformed into AW91 (clb5::URA3 clb6) and selected on 5-FOA. Correct clb5::CLB4 clb6 integrants were identified by PCR analysis.
2D agarose gel electrophoresis and analysis of replication fork direction. These techniques were carried out as described in Donaldson et al. (1998).
Cell synchronization, western blotting and flow cytometry. All strains were bar1. Cells were synchronized with 200 nM α-factor and released by the addition of pronase (0.3 mg/ml) to the medium. Protein sample extraction and western blotting were based on Stirling et al. (1994) and standard techniques. Protein loading was checked by Ponceau S staining of blots, followed by detection of Clb4-HA using antibody 12CA5. Flow cytometry was as described by Haase and Lew (1997).
Acknowledgments
ACKNOWLEDGEMENTS
I owe a great debt to Julian Blow for his endless encouragement, suggestions and enthusiasm, and for providing lab space. Thanks to Fred Cross for communicating data prior to publication and for strains. I am grateful to Mike Stark and members of the Dundee Yeast Laboratory for discussion and technical advice. Julian Blow and Mike Stark made useful comments on the manuscript. This work was funded by the Cancer Research Campaign (grant SP2385/0101) and the Grant Simpson Trust. I am a Royal Society University Research Fellow.
REFERENCES
- Amon A., Irniger, S. and Nasmyth, K. (1994) Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell, 77, 1037–1050. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman, W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–71. [DOI] [PubMed] [Google Scholar]
- Cross F.R., Yuste-Rojas, M., Gray, S. and Jacobson, M.D. (1999) Specialization and targeting of B-type cyclins. Mol. Cell, 4, 11–19. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Raghuraman, M.K., Friedman, K.L., Cross, F.R., Brewer, B.J. and Fangman, W.L. (1998) CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell, 2, 173–182. [DOI] [PubMed] [Google Scholar]
- Epstein C.B. and Cross, F.R. (1992) CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev., 6, 1695–1706. [DOI] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer, B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol., 262, 613–627. [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Diller, J.D., Ferguson, B.M., Nyland, S.V.M., Brewer, B.J. and Fangman, W.L. (1996) Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev., 10, 1595–1607. [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Brewer, B.J. and Fangman, W.L. (1997) Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells, 2, 667–678. [DOI] [PubMed] [Google Scholar]
- Haase S.B. and Lew, D.J. (1997) Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol., 283, 322–332. [DOI] [PubMed] [Google Scholar]
- Haase S.B. and Reed, S.I. (1999) Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature, 401, 394–396. [DOI] [PubMed] [Google Scholar]
- Kühne C. and Linder, P. (1993) A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J., 12, 3437–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini, D.J., Shah, N.J., Wach, A., Brachat, A., Philippsen, P. and Pringle, J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (1996) At the heart of the budding yeast cell cycle. Trends Genet., 12, 405–412. [DOI] [PubMed] [Google Scholar]
- Richardson H., Lew, D.J., Henze, M., Sugimoto, K. and Reed, S.I. (1992) Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev., 6, 2021–2034. [DOI] [PubMed] [Google Scholar]
- Schwob E. and Nasmyth, K. (1993) CLB5 and CLB6, a new pair of B-cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev., 7, 1160–1175. [DOI] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D. and Futcher, B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling D.A., Welch, K.A. and Stark, M.J. (1994) Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO J., 13, 4329–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]