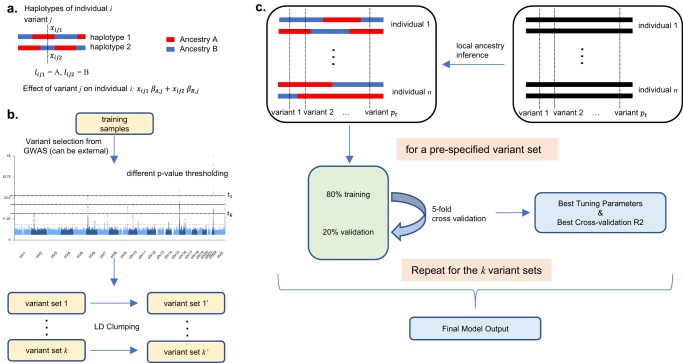
Fig. 1. Overview of GAUDI model and framework.
a Model set-up of GAUDI. Consider the haplotypes of individual i at variant j and assume local ancestry is already inferred. We consider the scenario with only two ancestries, namely A and B. Let denote haplotype value (taking values 0 or 1 for a directly genotyped variant, and ranging from 0 to 1 for an imputed variant). Let denote the local ancestry; here we have . Let denote population A, B specific effect of variant j on the phenotype. Thus we have the total effect of variant j in individual i as . b Variant selection framework of GAUDI. We first perform GWAS or use external GWAS results to obtain p-values, which will be used for variant selection. Specifically, we use the thresholding strategy to identify variants that are marginally associated with the trait of interest at k pre-specified p-value thresholds, . These k sets of variants will be generated, and we then perform LD clumping for each of the k sets to both reduce dimension and remove variants in high LD. c Final PRS construction of GAUDI. After inferring the local ancestry for every participant in the training set, for a specific set of variants, we perform five-fold cross-validation to select the best tuning parameters, under the penalized regression framework. Repeating the process for the k variant sets and comparing the cross-validated R2 will give us the final PRS model.