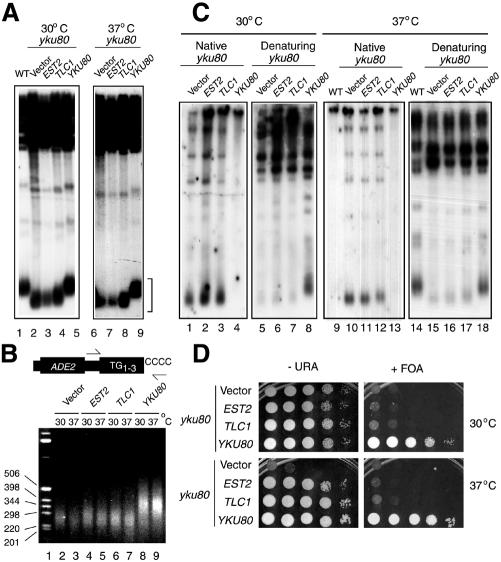
Fig. 1. (A) Southern blot of XhoI-digested genomic DNA probed with Y′-specific sequences of yeast telomeres. Genomic DNA was prepared from strains grown at 30°C (left) or 37°C (right) for 18 h, from wild-type (lane 1) or yku80 mutant strains containing either the YEP13 (2µ) plasmid (lanes 2, 6), YEP13–EST2 (lanes 3, 7), –TLC1 (lanes 4, 8) or –YKU80 (lanes 5, 9). Identical results (with the exception of yku80 with YEP13) were obtained with strains grown for >100 generations. Telomeric restriction fragments are indicated by a square bracket. (B) Telomere PCR. Genomic DNA was isolated, denatured and tailed with dCTP using TdT. The G-rich strand is specifically amplified with a primer complementary to the dC tail and a primer specific for a sequence distal of the subtelomeric ADE2 gene. The bottom panel shows PCR products analysed on an ethidium bromide-stained agarose gel. (C) In-gel hybridization of XhoI-digested genomic DNA probed with a radiolabelled oligonucleotide corresponding to the CA-repeat sequence of yeast telomeres, in native or denaturing conditions. Quantitation of the single-strand telomeric regions in yku80 mutants with or without EST2 or TLC1 overexpression showed no reproducible differences. (D) TPE was tested using a strain containing a telomere-proximal URA3 gene. Yeast strains were spotted in 10-fold serial dilutions on plates with (right) or without (left) FOA, and grown at 30°C (top) or 37°C (bottom) for 3 days.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.