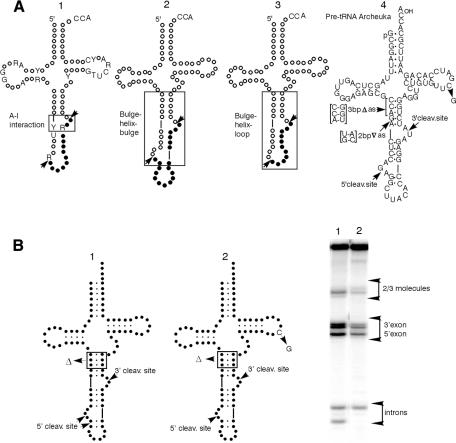
Fig. 1. (A) The A-I interaction. A conserved purine residue in the intron three nucleotides from the 3′ cleavage site (molecule 1, R in box) must pair with a pyrimidine in the anticodon loop 6 nucleotides upstream of the 5′ cleavage site (molecule 1, Y in box) to form the A-I (for anticodon–intron) interaction (Baldi et al., 1992). BHB (molecule 2). Two bulges of three nucleotides each (where cleavage occurs) rigidly separated by four base pairs (Daniels et al., 1985; Diener and Moore, 1998). BHL (molecule 3). A three-nucleotide 3′ site bulge, a four base-pair helix and a loop containing the 5′ site. Pre-tRNAArcheuka and its variants (molecule 4). The hybrid pre-tRNA molecule pre-tRNAArcheuka is a substrate for both the eukaryal and archaeal endonucleases. It consists of two regions derived from yeast pre-tRNAPhe [nucleotides (nt) 1–31 and nt 38–76] joined by a 25 nt insert that corresponds to the BHB motif of the archaeal pre-tRNATrp. It has a typical eukaryal mature domain with cleavage sites located at the prescribed distance from the reference elements and a correctly-positioned A-I base pair, all of which should ensure correct recognition by the eukaryal endonuclease when the enzyme operates in the mature-domain dependent mode. In addition, the presence of the BHB motif confers substrate characteristics that are recognizable by the eukaryal enzyme when it operates in the mature-domain independent mode. (B) A substrate cleaved in both the mature-domain dependent and the mature domain independent modes. Products of digestion by the Xenopus tRNA splicing endonuclease. Molecule 1, pre-tRNAArcheuka 3bpΔas; molecule 2, pre-tRNAArcheuka 3bpΔas, C56G.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.