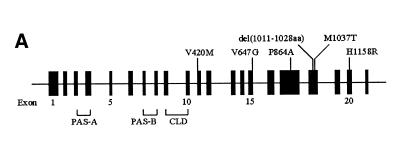
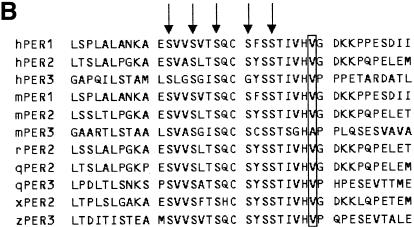
Fig. 1. (A) Schematic diagram of the hPer3 gene illustrating the locations of six polymorphisms with putative amino acid changes. Exons 1–21 are depicted as closed boxes and are numbered intermittently for clarity. Sizes of the introns and exons are not to scale. Polymorphisms with putative amino acid changes are shown above the gene. At the bottom are summarized the locations of the coding sequences for domains of hPER3. CLD, cytoplasmic localization domain. (B) Alignment of the amino acid sequences of vertebrate PER homologs. Only the region adjacent to the V647G polymorphism is shown. Amino acid residues that correspond to V647 in hPER3 are boxed. Arrows indicate the putative target residues of CK I ɛ (Toh et al., 2001). hPER1/2/3, human PER1/2/3; mPER1/2/3, mouse PER1/2/3; rPER2, rat PER2; qPER2/3, Japanese quail PER2/3; xPER2, Xenopus PER2; zPER3, zebrafish PER3.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.