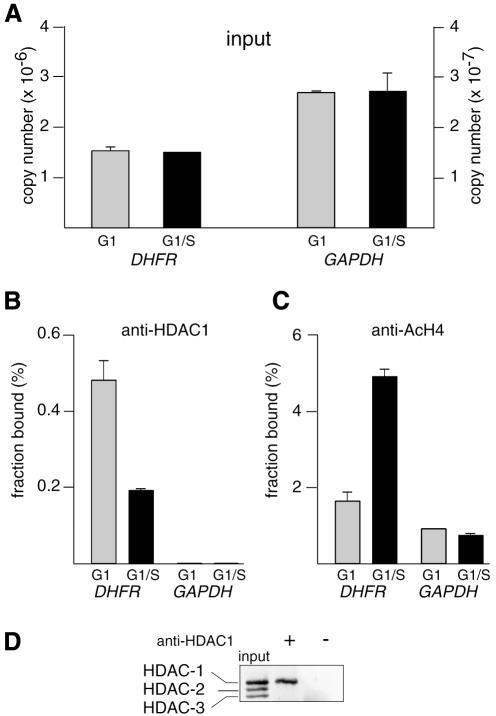
Fig. 2. Cell cycle-dependent recruitment of HDAC-1 and histone H4 deacetylation on DHFR promoter: compilation of results from Figure 1. Chromatin from synchronized NIH 3T3 cells, either in G1 phase (gray bars) or at the G1–S transition (black bars), was analyzed by the XChIP procedure, followed by quantitative PCR of eluted DNA. Equal amounts of chromatin (A) were subjected to immunoprecipitation using anti-HDAC-1 (B) or anti-AcH4 (C) antibodies. DHFR or GAPDH sequences (as indicated) were detected by quantitative PCR. Copy numbers were estimated by reference to a plasmid containing the promoter sequence and used as a standard; (A) number of copies in the inputs; (B) and (C) fraction of the total number of copies detected as antibody-bound material; a control sample run in parallel in the absence of antibodies revealed little association of DHFR (0.01% of the input) or GAPDH (0.002% of the input). Shown are the results of a typical experiment, with range bars indicating standard deviations of triplicates. This experiment has been reproduced four times with similar results. (D) Chromatin, immunoprecipitated by anti-HDAC-1 (+) or control (–) antibodies or before immunoprecipitation (input), was incubated for 15 min at 95°C and analyzed by western blotting, using an antibody that recognizes HDAC-1, HDAC-2 and HDAC-3 (H80920; Transduction Laboratories).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.