Abstract
Reconstitution of telomerase activity is proposed as a potential gene therapy to prevent, or rescue, age-related diseases produced by critical telomere shortening. However, it is not known whether or not short telomeres are irreversibly damaged. We addressed this by re-introducing telomerase in late generation telomerase-deficient mice, Terc–/–, which have short telomeres and show severe proliferative defects. For this, we have crossed these mice with Terc+/– mice and analyzed telomere length, chromosomal instability and premature aging of the progeny. The Terc–/– progeny had one set of chromosomes with normal telomeres, whereas the other set remained with critically short telomeres; these mice presented chromosomal instability and premature aging. In contrast, Terc+/– progeny showed all chromosomes with detectable telomeres, and did not show chromosomal instability or premature aging. These results prove that critically short telomeres can be rescued by telomerase, and become fully functional, thus rescuing premature aging. This has important implications for the future design of telomerase-based gene therapy of age-related diseases.
INTRODUCTION
Telomeres are DNA–protein complexes at the ends of eukaryotic chromosomes that protect them from degradation, recombination and DNA repair activities (reviewed in Blackburn, 1991, 2000; Greider, 1996). Loss of telomeric function by loss of telomeric repeats (TTAGGG in all vertebrates), or by mutation of telomere-binding proteins (i.e. TRF2, Ku proteins, DNA-PKcs), results in increased chromosomal instability both in cultured cells and in mice (Counter et al., 1992; Blasco et al., 1997; van Steensel et al., 1998; Hsu et al., 2000; Samper et al., 2000; Goytisolo et al., 2001).
Telomerase, the cellular reverse transcriptase (Tert, telomerase reverse transcriptase) that elongates telomeres de novo using an associated RNA molecule (Terc, telomerase RNA component) as template (Greider and Blackburn, 1985; reviewed in Nugent and Lundblad, 1998), has been at the spotlight of biomedicine due to its potential use in designing gene therapies for both cancer and aging. On one hand, telomerase activity is upregulated in the vast majority of human tumors compared with normal somatic tissues (reviewed in Shay and Bacchetti, 1997), and its inhibition in human tumor cell lines leads to telomere shortening and loss of cell viability (Hahn et al., 1999; Zhang et al., 1999), suggesting that telomerase inhibition could be an effective way to abolish tumor growth (reviewed in Zumstein and Lundblad, 1999). In support of this, mice that lack telomerase and have critically short telomeres display severe proliferative defects and, in the presence of wild-type p53, they are less susceptible to developing tumors (Chin et al., 1999; Greenberg et al., 1999; González-Suárez et al., 2000). Conversely, re-introduction of telomerase activity in cultured human primary cells results in telomere maintenance and immortal growth (Bodnar et al., 1998; Jiang et al., 1999; Morales et al., 1999; Ramirez et al., 2001).
Late generation telomerase-deficient mice, Terc–/–, have short telomeres and show defects in various proliferative tissues (Blasco et al., 1997; Lee et al., 1998; Herrera et al., 1999a,b, 2000; Rudolph et al., 1999). We show here that re-introduction of one copy of the Terc gene in these mice via Terc+/– × Terc–/– crosses is sufficient to elongate critically short telomeres, rescue chromosomal instability and prevent severe proliferative defects in these mice. These data clearly establish that the pathologies described in late generation Terc–/– mice are due to telomerase deficiency and telomere shortening and not to secondary mutations. These results also support the notion that telomerase activity re-introduction in adult somatic cells or tissues could be a potential approach for gene therapy of those age-related diseases triggered by telomere exhaustion, or for premature aging syndromes characterized by a faster rate of telomere shortening such as the Werner syndrome or Dyskeratosis congenita (Mitchell et al., 1999; Wyllie et al., 2000).
RESULTS AND DISCUSSION
Germline Terc re-introduction in late generation C57Bl6 Terc–/– mice via Terc–/– × Terc+/– crosses
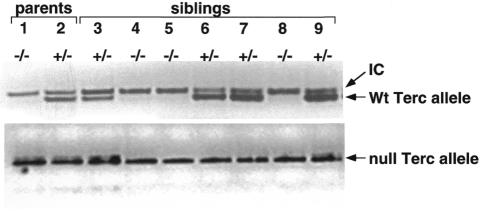
Mice deficient for the Terc component of telomerase, Terc–/–, lack telomerase activity and show telomere shortening and chromosomal instability with increasing mouse generations until infertility occurs at the fourth (G4) or sixth (G6) generation depending on the genetic background (G4 for mice in a pure C57Bl6 background; G6 for mice in a mixed C57Bl6/129Sv/SJL background) (Blasco et al., 1997; Lee et al., 1998; Herrera et al., 1999b; Rudolph et al., 1999). In contrast to late generation G6 Terc–/– in the mixed background (Rudolph et al., 1999), late generation G4 Terc–/– mice in the C57Bl6 background show severe phenotypes at very young ages (1–5 months of age) (Herrera et al., 1999b; this paper). To study whether re-introduction of the Terc gene in the germline of these mice is sufficient to prevent proliferative defects associated to telomere shortening, we studied the progeny of G3 Terc–/– × Terc+/– crosses. The G4 progeny resulting from this cross will have a set of chromosomes with short telomeres from the G3 Terc–/– parent and a set of chromosomes with normal telomeres from the Terc+/– parent. However, only the G4 Terc+/– progeny will inherit a copy of the Terc gene and have telomerase activity reconstitution. We have focused our study on the full characterization of the parents and the progeny of a male G3 Terc–/– (number 1) × female Terc+/– (number 2) cross (Figure 1). Seven different littermates were obtained, three of them were G4 Terc–/– (numbers 4, 5 and 8 in Figure 1) and four of them were G4 Terc+/– (numbers 3, 6, 7 and 9 in Figure 1). Figure 1 shows the presence or absence of the wild-type Terc allele in the parents and the progeny using a PCR-based method to detect the wild-type and null Terc alleles (see Methods).
Fig. 1. Generation of G4 Terc+/– and G4 Terc–/– littermates via a G3 Terc–/– × Terc+/– cross. Wild-type and null Terc alleles were identified by PCR (see Methods). The positions of the PCR bands corresponding to the wild-type and null alleles, as well as that of an internal control (IC) for PCR efficiency, are indicated by arrows.
Re-introduction of the Terc gene is sufficient to restore short telomeres to their normal length in G4 mice
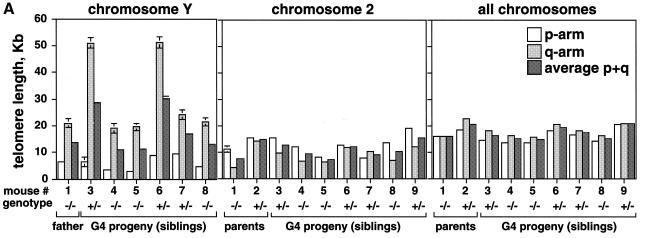
We addressed whether the G4 progeny that received the Terc wild-type allele rescued the critically short telomeres coming from the G3 Terc–/– father. For this, we first determined telomere length of the ‘Y’ chromosome in the G3 Terc–/– father and in the male G4 progeny using quantitative telomeric FISH on primary splenocytes (Q-FISH; Zijlmans et al., 1997; Samper et al., 2000). Table I and Figure 2A show that both p- and q-Y-telomeres were elongated from 6.4 ± 0.4 kb (p-arm) and 21.0 ± 1.6 kb (q-arm) in the father to an average length of 8.8 ± 0.5 kb (p-arm) and 42.1 ± 1.9 kb (q-arm) in the G4 Terc+/– siblings (average of mice 3, 6, 7 and 9 in Figure 2A and Table I). This corresponds to an average elongation of 2.0 and 21.1 kb for p- and q-telomeres, respectively. It is relevant to note that q-Y-telomeres were subjected to larger changes in length than p-Y-telomeres in the presence of the Terc gene, suggesting that they may be more ‘accessible’ to telomerase. Both p- and q-Y-telomeres shortened from 6.4 ± 0.4 kb (p-arm) and 21.0 ± 1.6 kb (q-arm) in the father to an average of 3.8 ± 0.3 kb (p-arm) and 20.1 ± 1.5 kb (q-arm) in the G4 Terc–/– siblings (average of mice 4, 5 and 8 in Figure 2A and Table I). This corresponds to an average shortening of 2.6 and 0.9 kb for p- and q-Y-telomeres, respectively, in agreement with the absence of telomerase activity in these mice. Altogether, these results indicate that telomerase activity is able to elongate short telomeres (i.e. those of the parental G3 Terc–/– Y-chromosome) extending them to different lengths in the G4 Terc+/– progeny.
Table I. Telomere length in parents and progeny from a Terc+/– × G3 Terc–/– cross.
| Parents |
Progeny (siblings) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Generation | G3 | G4 | G4 | G4 | G4 | G4 | G4 | G4 | |
| Genotype | –/– | +/– | +/– | –/– | –/– | +/– | +/– | –/– | +/– |
| Mouse number/sex | 1 (xy) | 2 (xx) | 3 (xy) | 4 (xy) | 5 (xy) | 6 (xy) | 7 (xy) | 8 (xy) | 9 (xx) |
| Q-FISHa | p: 6.4 ± 0.4 | p: 6.4 ± 0.4 | p: 3.6 ± 0.4 | p: 3.0 ± 0.3 | p: 9.0 ± 0.5 | p: 9.6 ± 0.7 | p: 4.8 ± 0.3 | ||
| Y-chromosomeb | q: 21.0 ± 1.6 | female | q: 51.0 ± 1.9 | q: 19.1 ± 1.7 | q: 19.6 ± 1.4 | q: 51.2 ± 2.0 | q: 24.2 ± 1.8 | q: 21.6 ± 1.5 | female |
| t: 13.7 ± 0.7 | t: 28.7 ± 0.8 | t: 11.4 ± 0.7 | t: 11.3 ± 0.6 | t: 30.1 ± 0.9 | t: 16.9 ± 0.8 | t: 13.2 ± 0.6 | |||
| Q-FISHa | p: 11.1 ± 1.2 | p: 15.5 ± 0.6 | p: 15.5 ± 0.5 | p: 12.0 ± 0.5 | p: 8.1 ± 0.9 | p: 12.5 ± 0.6 | p: 7.8 ± 0.3 | p: 13.5 ± 0.7 | p: 19.1 ± 0.7 |
| chromosome 2c | q: 4.1 ± 0.3 | q: 14.1 ± 0.6 | q: 9.5 ± 0.5 | q: 6.5 ± 0.6 | q: 6.1 ± 0.9 | q: 11.8 ± 0.7 | q: 10.3 ± 0.4 | q: 6.8 ± 0.6 | q: 11.9 ± 0.4 |
| t: 7.6 ± 0.9 | t: 14.8 ± 0.5 | t: 12.5 ± 0.5 | t: 9.2 ± 0.6 | t: 7.1 ± 0.8 | t: 12.1 ± 0.5 | t: 9.0 ± 0.4 | t: 10.1 ± 0.8 | t: 15.5 ± 0.7 | |
| Q-FISHa | p: 16.3 ± 0.3 | p: 18.6 ± 0.2 | p: 14.6 ± 0.2 | p: 13.8 ± 0.2 | p: 13.7 ± 0.3 | p: 18.2 ± 0.2 | p: 16.9 ± 0.3 | p: 14.3 ± 0.3 | p: 20.6 ± 0.3 |
| (all chromosomes)b | q: 16.1 ± 0.3 | q: 22.7 ± 0.3 | q: 18.3 ± 0.3 | q: 16.6 ± 0.3 | q: 16.0 ± 0.3 | q: 20.8 ± 0.3 | q: 18.2 ± 0.3 | q: 16.5 ± 0.2 | q: 21.1 ± 0.3 |
| t: 16.2 ± 0.3 | t: 20.7 ± 0.2 | t: 16.4 ± 0.2 | t: 15.2 ± 0.3 | t: 14.9 ± 0.3 | t: 19.5 ± 0.3 | t: 17.6 ± 0.3 | t: 15.4 ± 0.3 | t: 20.9 ± 0.3 | |
aData also shown in Figure 2A.
bOnly males are included.
cBoth males and females are included.
p, p-arm; q, q-arm; t, average of p- and q-arms.
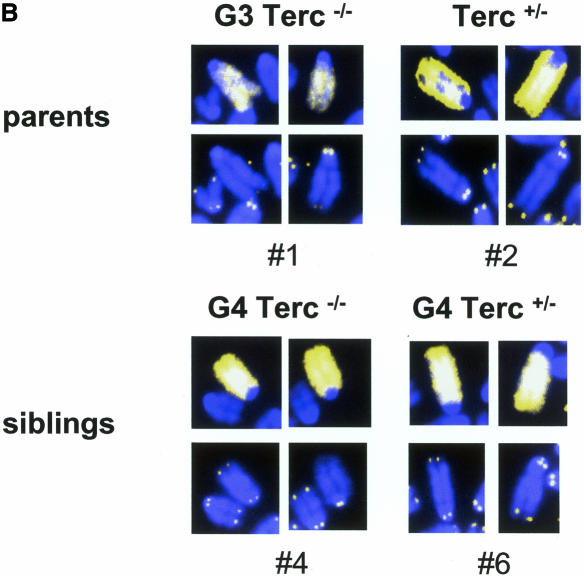
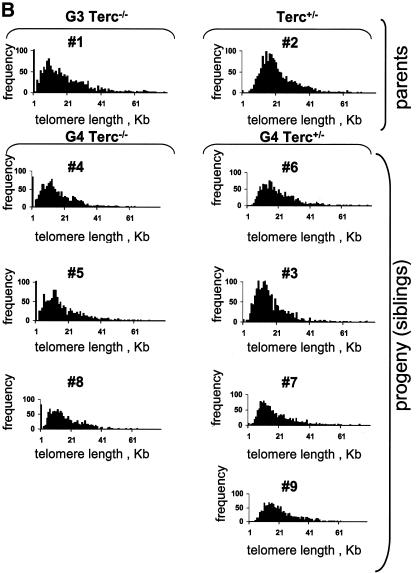
Fig. 2. (A) Telomere length of both p- and q-arms, as well as the average of both arms (average p+q) from chromosome Y, chromosome 2, as well as all chromosomes, are represented. Standard errors are also included; however, some of the errors are so small (see Table I) that the error bars are not visible. (B) Illustrative images of combined Q-FISH and chromosome 2 painting. For each case represented, both chromosome 2 are shown (mice numbers 1, 2, 4 and 6). Notice undetectable chr2-telomeres in the G3 Terc–/– father (number 1), as well as in the G4 Terc–/– sibling (number 4). The G4 Terc+/– sibling (number 6) and the Terc+/– mother (number 2) show detectable chr2-telomeres.
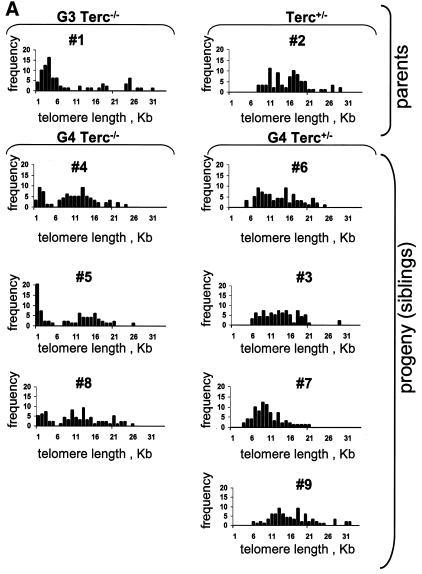
We also determined the length of chromosome 2 telomeres (chr2-telomeres) in the parents and the progeny using Q-FISH and a chr2-specific painting probe on primary splenocytes (Methods) (see Figure 2B for examples). Chromosome 2 was chosen for the study because, in the mouse, this chromosome has shorter telomeres than average (Hande et al., 1999). Chr2-telomeres (average of p- and q-arms) in the G3 Terc–/– father were shorter than in the Terc+/– mother, 7.6 ± 0.9 and 14.8 ± 0.5 kb, respectively (mice 1 and 2 in Table I and Figure 2A). Figure 2B shows that the G3 father has undetectable telomeres in one chromosome 2, whereas the Terc+/– mother has both copies of chromosome 2 with normal telomeres. This is also illustrated by telomere length histograms, which show that the G3 Terc–/– father has a population of very short chr2-telomeres, which are not present in the Terc+/– mother (compare numbers 1 and 2 in Figure 3A; see also Figure 2B for representative examples). In the case of the G4 progeny, chr2-telomeres were longer in the siblings that received the Terc allele than in those that received the null allele, 12.3 ± 0.5 and 8.8 ± 0.7 kb, respectively (average of telomere length values shown for individual mice in Table I and Figure 2A; see Figure 2B for representative examples). Importantly, histograms depicting chr2-telomere length frequencies revealed that only those siblings that received the wild-type Terc allele rescued the population of very short chr2-telomeres inherited from the father (G4 Terc+/–; numbers 3, 6, 7 and 9 in Figure 3A; see mouse number 6 in Figure 2B for a representative example), whereas those siblings that received the null allele showed a two-peak distribution of telomeres: a peak of very short telomeres (inherited from the father) and a peak of normal telomeres (inherited from the mother) (G4 Terc–/–; numbers 4, 5 and 8 in Figure 3A, see mouse number 4 in Figure 2B for a representative example). These observations indicate that telomerase is able to rescue short telomeres in a mouse, as previously shown for Y-telomeres.
Fig. 3. (A) Telomere length distribution of chr2-telomeres in primary splenocytes from the parents and the G4 progeny as determined by Q-FISH. The histogram depicts a population of critically short chr2-telomeres in the G3 father and in the G4 Terc–/– littermates. In contrast, the G4 Terc+/– littermates show disappearance of the peak corresponding to short telomeres. (B) Telomere length distribution of all chromosome telomeres in primary splenocytes from the parents and the G4 progeny as determined by Q-FISH. The histogram depicts a population of critically short telomeres (≤1 kb) in the G3 father and in the G4 Terc–/– littermates. The G4 Terc+/– littermates show disappearance of ≤1 kb telomeres.
Curiously, we noticed that chr2-q-telomeres were shorter than chr2-p-telomeres both in the parents and the progeny, suggesting that the general assumption that p-telomeres are shorter than q-telomeres is not always true (Zijlmans et al., 1997).
Q-FISH analysis of all-chromosome telomeres was also performed in the parents and the progeny. The average length of all telomeres was shorter in the G3 Terc–/– father than in the Terc+/– mother, 16.2 ± 0.3 and 20.7 ± 0.2 kb, respectively (Table I; Figure 2A). Similarly, those G4 siblings that received the wild-type Terc allele showed longer telomeres than the ones that received the null allele; average telomere lengths were 18.6 ± 0.2 and 15.6 ± 0.3 kb, respectively (Table I; Figure 2A). Histograms depicting telomere length frequencies also indicated that the G4 Terc+/– but not the G4 Terc–/– progeny rescued the population of critically short telomeres (notice telomeres ≤1 kb in the G4 Terc–/– but not the G4 Terc+/– progeny; Figure 3B). In agreement with this, all G4 Terc–/– siblings had a very high percentage of chromosome-ends with undetectable TTAGGG repeats by Q-FISH (∼6% of all ends lacked detectable TTAGGG signals) compared with the G4 Terc+/– siblings (only ∼0.2% of all ends lacked detectable TTAGGG signal) (Table II shows data for each individual mouse). Again, these results suggest that telomerase is sufficient to rescue short telomeres in a mouse, as shown above for Y- and chr2-telomeres.
Table II. Chromosomal aberrations in the parents and progeny of a G3 Terc–/– × Terc+/– cross.
| Mouse number and genotype | Chromosomal aberrations | Undetectable telomeres (%) |
|---|---|---|
| #1 G3 Terc–/– | 3.8 RTa | 204/1761b |
| 3.8 B/F | (11.5%) | |
| #2 Terc+/– | 0 RT | 6/1580 |
| 3.7 B/F | (0.3%) | |
| #3 G4 Terc+/– | 0 RT | 6/1568 |
| 0 B/F | (0.4%) | |
| #4 G4 Terc–/– | 11.1 RT | 83/1573 |
| 3.7 B/F | (5.3%) | |
| #5 G4 Terc–/– | 38.7 RT | 105/1272 |
| 3.2 DIC | (8.25%) | |
| 0 B/F | ||
| #6 G4 Terc+/– | 0 RT | 0/1600 |
| 0 B/F | (0.0%) | |
| #7 G4 Terc+/– | 0 RT | 5/1600 |
| 0 B/F | (0.3%) | |
| #8 G4 Terc–/– | 8 RT | 82/1596 |
| 12 B/F | (5.1%) | |
| #9 G4 Terc+/– | 3.4 RT | 0/1612 |
| 0 B/F | (0.0%) |
aAberrations per 100 metaphases.
bEnds lacking TTAGGG signal out of total ends analyzed. The percentage of undetectable telomeres is also shown in parentheses.
RT, Robertsonian-like chromosome; B/F, breaks/fragments; DIC, dicentric chromosome.
Curiously, Q-FISH analysis showed that Y-telomeres (especially Y-q-telomeres) were longer than the average of all chromosome telomeres, whereas those of chromosome 2 were shorter than average (Figure 1A).
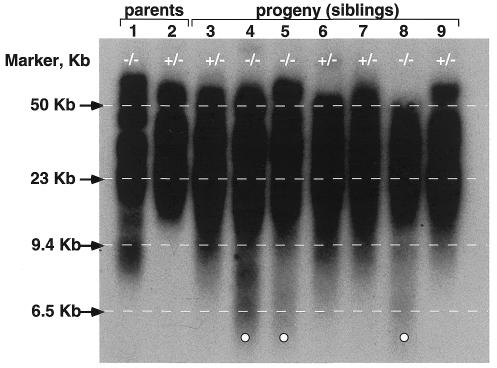
As a different approach to estimate telomere length that is not based on fluorescence, we carried out terminal restriction fragment (TRF) analysis using Southern blotting (Methods). Figure 4 shows TRF analysis of both parents, as well as that of the progeny. First, it is noticeable that the G3 Terc–/– father shows low molecular weight TRFs (below the 9.4 kb marker), which are not present in the Terc+/– mother (Figure 4). All siblings that received the wild-type Terc allele from the Terc+/– mother, G4 Terc+/– (numbers 3, 6, 7 and 9), showed low molecular weight TRFs of ≥9.4 kb, whereas siblings that received the null Terc allele from the mother, G4 Terc–/– (numbers 4, 5 and 8), showed low molecular weight TRFs that were below the 6.5 kb marker (also indicated by white circles in Figure 4). These observations suggest that the appearance of low molecular weight TRF bands correlates with telomere shortening in the G4 progeny, and that G4 Terc–/– mice have shorter telomeres than the G4 Terc+/– littermates. The TRF analysis is also in agreement with the notion that short telomeres are elongated by telomerase when the Terc wild-type allele is present, as previously shown by Q-FISH.
Fig. 4. Measurement of terminal restriction fragments (TRFs) in bone marrow cells from both parents and the G4 progeny. Decrease in telomere length is visualized by the appearance of TRF bands below the 6.5 kb marker in the G4 Terc–/– mice that are not present in the G4 Terc+/– littermates (indicated by open circles). +/–, Terc+/– (numbers 2, 3, 6, 7 and 9); –/–, Terc–/– (numbers 1, 4, 5 and 8).
Rescue of chromosomal instability in G4 Terc+/– siblings
Critically short telomeres in late generation Terc–/– mice in two different genetic backgrounds result in telomere dysfunction and increased chromosomal instability, mostly consisting of end-to-end fusions (Robertsonian-like fusions and dicentrics) (Blasco et al., 1997; Herrera et al., 1999b). Here, we studied whether re-introduction of the Terc gene in these mice, hence of telomerase activity, is able to rescue chromosomal instability. For this, we scored chromosomal aberrations on primary splenocytes using Q-FISH. Table II shows the frequencies of different chromosomal aberrations in the parents and the G4 progeny. All G4 siblings that received the null allelle, G4 Terc–/– (numbers 4, 5 and 8 in Table II), showed increased end-to-end fusions, as well as chromosome breaks and fragments compared with the G4 Terc+/– littermates (numbers 3, 6, 7 and 9 in Table II) and with the Terc+/– mother (number 2 in Table II). As an exception, G4 Terc+/– mouse number 9 showed a low frequency of RT fusions; however, it did not show chromosome ends without detectable TTAGGG repeats (Table II), suggesting that these fusions were not due to critically short telomeres and may have been inherited directly from the father. The G4 Terc–/– progeny also showed a higher chromosomal instability than the G3 Terc–/– father (number 1 in Table II), in agreement with further telomere shortening in the absence of telomerase (see above).
Telomerase rescues premature aging phenotypes in G4 Terc+/– mice
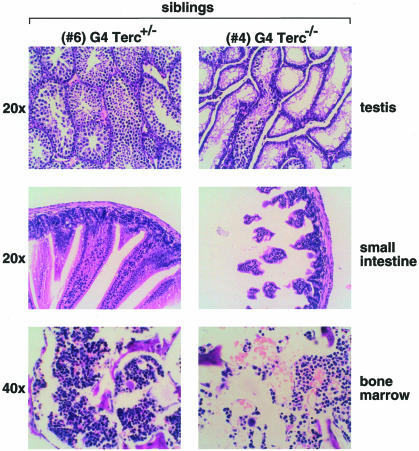
To study whether telomerase-mediated rescue of short telomeres and of chromosomal instability also prevented the occurrence of severe proliferative phenotypes in the Terc–/– mice, we did a complete histopathological analysis of the parents and the G4 progeny 21 days post-partum (Table III). All G4 Terc–/– siblings (numbers 4, 5 and 8) showed bone marrow aplasia, spleens dramatically reduced in size (not shown), as well as different degrees of intestinal atrophy (see Figure 5 for example). As an exception, G4 Terc+/– mouse number 3 also showed reduced spleen size; however, this could be due to the fact that the G4 mice were only 21 days old at the time of the analysis and there could be variability in spleen size at early ages. One of the G4 Terc–/– males, number 4, showed a very severe testicular atrophy (Figure 5). These disease states are typical of late generation telomerase knockout mice in a C57Bl6 genetic background and have been associated with premature aging in these mice (Herrera et al., 1999b). In marked contrast, G4 Terc+/– littermates showed no pathologies, with the exception of a slight villi atrophy in mice numbers 3 and 6 (Table III). Figure 5 shows a comparison of three different tissues in two G4 littermates that carry, or not, a copy of the Terc gene (mice numbers 6 and 4, respectively). The rescue of severe premature aging phenotypes in the G4 progeny that received the Terc wild-type allele is in agreement with the previous data, which showed that all telomeres were restored to their normal length and chromosomal aberrations were prevented in these mice. However, those G4 mice that received the null Terc allele showed the characteristic severe disease states associated with short telomeres, in agreement with the fact that they carried a set of telomeres that were critically short (see Tables I and II, as well as Figure 3).
Table III. Phenotypes in littermate mice generated by Terc+/– × G3 Terc–/– (C57Bl6) intercrosses.
| Parents |
Progeny |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Generation | G3 | G3 | G4 | G4a | G4 | G4a | G4 | G4 | G4 |
| Genotype | –/– | +/– | +/– | –/– | –/– | +/– | +/– | –/– | +/– |
| Mouse number/sex | 1 (xy) | 2 (xx) | 3 (xy) | 4 (xy) | 5 (xy) | 6 (xy) | 7 (xy) | 8 (xy) | 9 (xx) |
| Testis | normal | – | normal | severe testicular atrophy | normal | normal | normal | normal | – |
| GI tract | slight villi atrophy in small intestine | normal | slight villi atrophy in small intestine | severe atrophy in small intestine | moderate atrophy in small intestine | slight villi atrophy in small intestine | normal | normal | normal |
| Bone marrow | normal | normal | normal | severe aplasia lymphoid cells | aplasia | normal | normal | aplasia | normal |
| Spleen | normal | normal | reduced size | reduced size | reduced size | normal | normal | reduced size | normal |
aImages of the pathologies observed in these mice are shown in Figure 5.
Fig. 5. Histology of testis, small intestine and bone marrow from littermate 21-day-old G4 Terc–/– and G4 Terc+/– siblings (numbers 4 and 6, respectively). Magnifications were 20 and 40×, as indicated.
Conclusions
Here we show that re-introduction of telomerase activity in late generation Terc–/– mice is sufficient to elongate short telomeres, rescue chromosomal instability and prevent to a large extent the severe phenotypes associated with telomere shortening in these mice. These observations have important implications for the future design of telomerase-based gene therapy of age-related diseases. In particular, we show here that (i) telomerase can elongate short telomeres in a mammalian organism and that (ii) this restoration of short telomeres by telomerase is sufficient to rescue chromosomal instability and to prevent the severe premature aging phenotypes associated with critically short telomeres. From these studies one can speculate that diseases which relate to defects in telomerase function such as Dyskeratosis congenita (Mitchell et al., 1999) might be treatable by restoring functional telomerase in stem cells of the affected individuals. Telomerase-based therapy can also be useful in patients who suffer from diseases characterized by a faster rate of telomere shortening, such as Werner syndrome patients, whose fibroblasts have recently been shown to recover proliferative capacity upon Tert over-expression (Wyllie et al., 2000). Furthermore, from the results presented here one might extrapolate that a putative anticancer therapy based on transient telomerase inhibition in cancer patients should not lead to irreversible side-effects in highly proliferative tissues, since the recovery of telomerase activity in these tissues will rescue critically short telomeres and prevent chromosomal damage and loss of proliferative capacity.
METHODS
Generation and genotyping of G4 Terc–/– and G4 Terc+/– littermates. Terc–/– mice in the C57BL6 background (Bl6) were described elsewhere (Herrera et al., 1999b). A 3-month-old Terc–/– male was crossed with a 6-month-old Terc+/– female to derive a litter of seven pups that were killed at 21 days post-partum due to the ill health of some of the littermates. Both parents were killed in parallel with the seven pups. A tail necropsy from all animals was taken to extract DNA for genotyping purposes. Germline identification of wild-type and null Terc alleles was carried out using both Southern blot genotyping (Blasco et al., 1997) and PCR genotyping (Argilla and Hanahan, unpublished data).
Mice handling. Mice were housed at our barrier area, where pathogen-free procedures are employed in all mouse rooms. Quarterly health monitoring reports have been negative for all pathogens in accordance with FELASA recommendations (Federation of European Laboratory Animal Science Associations).
Histopathological analyses. Testis, bone marrow and small intestine sections from the parents and the progeny (21 days post-partum) were fixed in 10% buffered formalin and hematoxylin-eosin stained. Images were captured with an Olympus-Vanox microscope at 20 or 40× magnification, as indicated.
Telomere length measurements. Q-FISH spleens were removed, washed in sterile phosphate-buffered saline (PBS) and ground between two pieces of nylon mesh to obtain fresh splenocytes. After lysis with 0.75% ammonium chloride, splenocytes were counted and cell cultures with 3–5 million cells were initiated in RPMI 1640 with 20% fetal calf serum, antibiotics, 20 µg/ml lipopolysaccharide and 5 µg/ml Con A. After 70 h in culture, 0.1 µg/ml colcemid was added for 2 h and metaphase cells were prepared by standard methods. Splenocyte metaphases were hybridized with a PNA-tel probe, and telomere length was determined as described (Zijlmans et al., 1997; Samper et al., 2000) using the TFL-TELO program (gift of Dr Lansdorp, Vancouver, Canada). The location of the metaphases was recorded for further analysis with a chromosome 2 painting probe (see below). After capturing the images for Q-FISH analysis, the coverslips were removed with acetone and the slides were washed in PBS for 10 min. After increasing ethanol series, slides were air-dried and denatured in 70% formamide, 2× SSC at 65°C for 2 min. Following denaturation, slides were quenched in ice-cold 70% ethanol, dehydrated in increasing ethanol series and air-dried. The Cy3-labeled chromosome 2 painting probe was obtained from Cambio Ltd (UK). Fifteen microliters of painting probe per slide were denatured for 10 min at 65°C and re-annealed at 37°C for 1.5 h, and then hybridized to the metaphases at 42°C overnight. Subsequently, slides were washed twice with 50% formamide in 2× SSC at 45°C for 5 min, and in 0.1× SSC at 45°C for 5 min. Finally, slides were mounted in Vectashield medium (Vector labs) with DAPI. Those same metaphases that were previously recorded for Q-FISH analysis were relocated, and the chromosome 2-hybridized metaphases were captured again. Fifteen different metaphases were analyzed in each case. For chromosomal instability determinations, between 25 and 31 metaphases were analyzed.
The chromosome Y was identified by reverse DAPI banding in the original image taken for Q-FISH.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Serrano and E. Santos for mice handling and genotyping. E.S. is a predoctoral fellow of the Regional Government of Madrid (CAM). Research at the laboratory of M.A.B. is funded by Swiss Bridge Award 2000, by grants PM97-0133 from the Ministry of Science and Technology (MCT), Spain, 08.1/0030/98 from the CAM, grants FIGH-CT1999-00009, FIGH-CT-1999-00002 and QLG1-1999-01341 from the EU, and by the Department of Immunology and Oncology (DIO). The DIO was founded and is supported by the Spanish Research Council (CSIC) and by the Pharmacia Corporation.
REFERENCES
- Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2000) Telomerase states and cell fates. Nature, 408, 53–56. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee, H.-W., Hande, P., Samper, E., Lansdorp, P., DePinho, R. and Greider, C.W. (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Bodnar A.G. et al. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- Chin L., Artandi, S.E., Shen, Q., Tam, A., Lee, S.L., Gottlieb, G.J., Greider, C.W. and DePinho, R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Counter C.M., Avilion, A.A., LeFeuvre, C.E., Stewart, N.G., Greider, C.W., Harley, C.B. and Bacchetti, S. (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J., 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Suárez E., Samper, E., Flores, J.M. and Blasco, M.A. (2000) Telomerase-deficient mice with short telomeres are resistant to skin tumorigenesis. Nature Genet., 26, 114–117. [DOI] [PubMed] [Google Scholar]
- Goytisolo F., Samper, E., Edmonson, S., Taccioli, G.E. and Blasco, M.A. (2001) Absence of DNA-PKcs in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol., 21, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R.A., Allsopp, R.C., Chin, L., Morin, G. and DePinho, R. (1999) Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell, 97, 515–525. [DOI] [PubMed] [Google Scholar]
- Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn, E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- Hahn W.C. et al. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nature Med., 5, 1164–1170. [DOI] [PubMed] [Google Scholar]
- Hande P., Samper, E., Lansdorp, P. and Blasco, M.A. (1999) Telomere length dynamics in cultured cells from normal and telomerase null mice. J. Cell Biol., 144, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E., Samper, E. and Blasco, M.A. (1999a) Telomere shortening in mTR–/– embryos is associated with failure to close the neural tube. EMBO J., 18, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E., Samper, E., Martín-Caballero, J., Flores, J.M., Lee, H.-W. and Blasco, M.A. (1999b) Disease states associated to telomerase deficiency appear earlier in mice with short telomeres. EMBO J., 18, 2950–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E., Martinez, A.C. and Blasco, M.A. (2000) Impaired germinal center reaction in mice with short telomeres. EMBO J., 19, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-L. et al. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev., 14, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.-R. et al. (1999) Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nature Genet., 21, 111–114. [DOI] [PubMed] [Google Scholar]
- Lee H.-W., Blasco, M.A., Gottlieb, G.J., Horner, J.W., Greider, C.W. and DePinho, R.A. (1998) Essential role of mouse telomerase in highly proliferative organs. Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Wood, E. and Collins, K. (1999) A telomerase component is defective in the human disease Dyskeratosis congenital. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- Morales C.P., Holt, S.E., Ouellette, M., Kaur, K.J., Yan, Y., Wilson, K.S., White, M.A., Wright, W.E. and Shay, J.W. (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nature Genet., 21, 115–118. [DOI] [PubMed] [Google Scholar]
- Nugent C.I. and Lundblad, V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Ramirez R.D., Morales, C.P., Herbert, B.S., Rohde, J.M., Passons, C., Shay, J.W. and Wright, W.E. (2001) Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev., 15, 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph K.L., Chang, S., Lee, H.-W., Blasco, M.A., Gottlieb, G., Greider, W.C. and DePinho, R.A. (1999) Longevity, stress response, and cancer in aging telomerase deficient mice. Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- Samper E., Goytisolo, F., Slijepcevic, P., van Buul, P. and Blasco, M.A. (2000) Mammalian Ku86 prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep., 1, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J.W. and Bacchetti, S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska, A. and de Lange, T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Wyllie F.S., Jones, C.J., Skinner, J.W., Haughton, M.F., Wallis, C., Wynford-Thomas, D., Faragher, R.G. and Kipling, D. (2000) Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nature Genet., 24, 16–17. [DOI] [PubMed] [Google Scholar]
- Zhang X., Mar, V., Zhou, W., Harrington, L. and Robinson, M.O. (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev., 13, 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans J.M., Martens, U.M., Poon, S., Raap, A.K., Tanke, H.J., Ward, R.K. and Lansdorp, P.M. (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA, 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumstein L.A. and Lundblad, V. (1999) Telomeres: has cancer’s Achilles’ heel been exposed? Nature Med., 5, 1129–1130. [DOI] [PubMed] [Google Scholar]