Figure 3. Hypoxia approximating physioxia increases HIF1A and ARNT binding to cis-regulatory elements of the Nos1 locus.
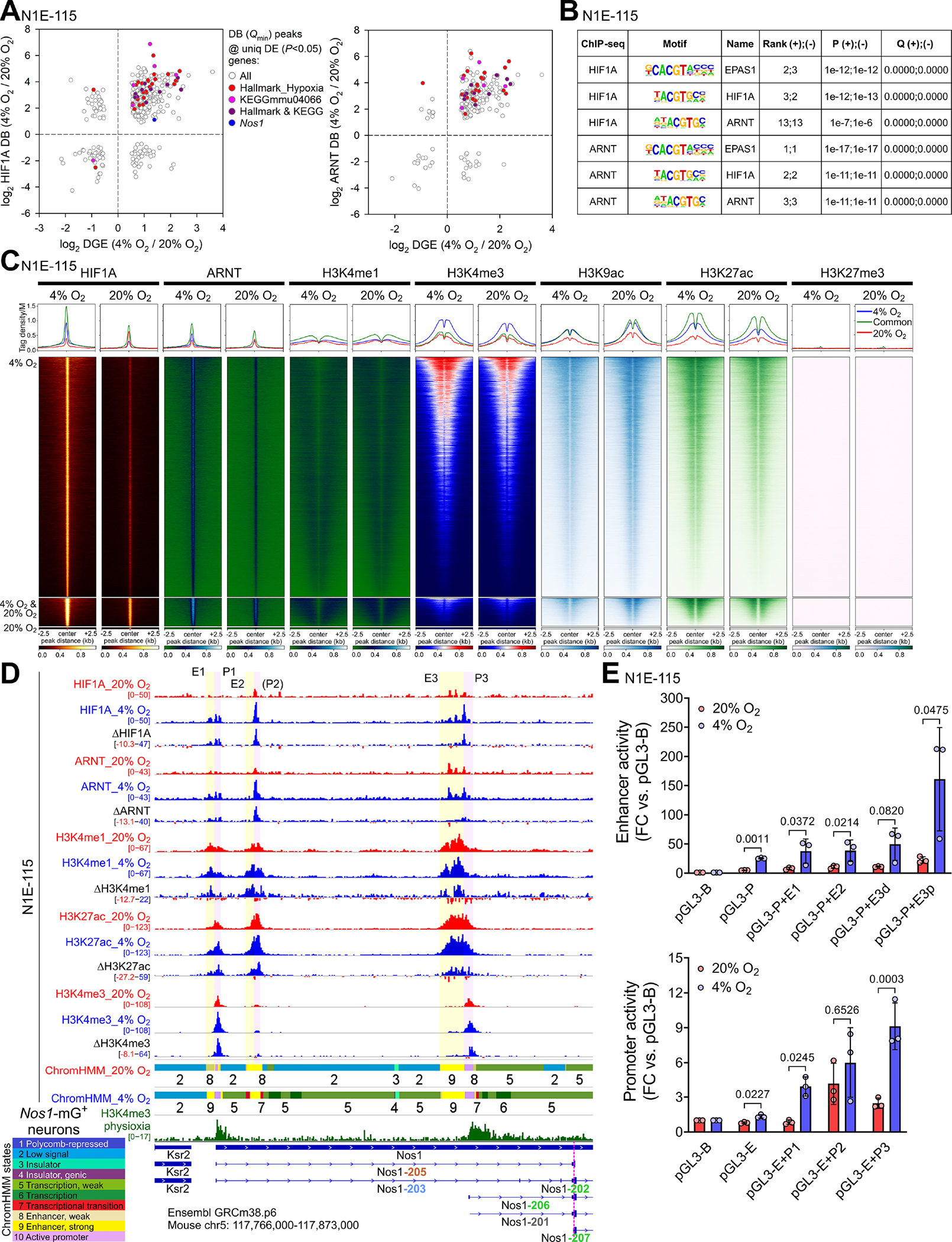
(A) Correlation between differential binding (DB; FDR Q<0.1) of HIF1A and ARNT within peaks with the lowest FDR Q value among the peaks assigned to the same gene and differential gene expression (DE; P<0.05) in N1E-115 cells cultured for 3 days at 4% or 20% O2. (B) Motifs within the HIF1A and ARNT peaks assigned to KEGG mmu04066 genes. (C) Heatmaps and average tag density plots of ChIP-seq data aligned to the centers of HIF1A peaks±2.5 kb are shown for HIF1A peaks unique to cells cultured at 4% O2 (blue lines, top heatmap panels) or 20% O2 (red lines, bottom heatmap panels) or common to cells cultured at 4% or 20% O2 (green lines, middle heatmap panels). (D) Binding profiles of HIF1A, ARNT, and key histone marks in the Ensembl GRCm38.p6 Nos1 locus of N1E-115 cells cultured at 20% (red tracks) or 4% O2 (blue) for 3 days. Aggregate and subtracted (4% O2−20% O2) occupancy data from two replicates are displayed. Colored ribbons: epigenetic states discovered by ChromHMM (10-state model). Green track: H3K4me3 in FACS-isolated Nos1-mG+ enteric neurons. (E) Activities of the indicated candidate Nos1 cis-regulatory elements cloned into the pGL3-Promoter (pGL3-P; top) or the pGL3-Enhancer (pGL3-E; bottom) vectors vs. the pGL3-Basic (pGL3-B) plasmid. The E3 putative enhancer was separated into a Nos1-distal (E3d) and Nos1-proximal (E3p) fragment. P, ratio-paired t tests.
