Figure 4. HIF1A controls Nos1 transcription through multiple cis-regulatory elements of the Nos1 locus.
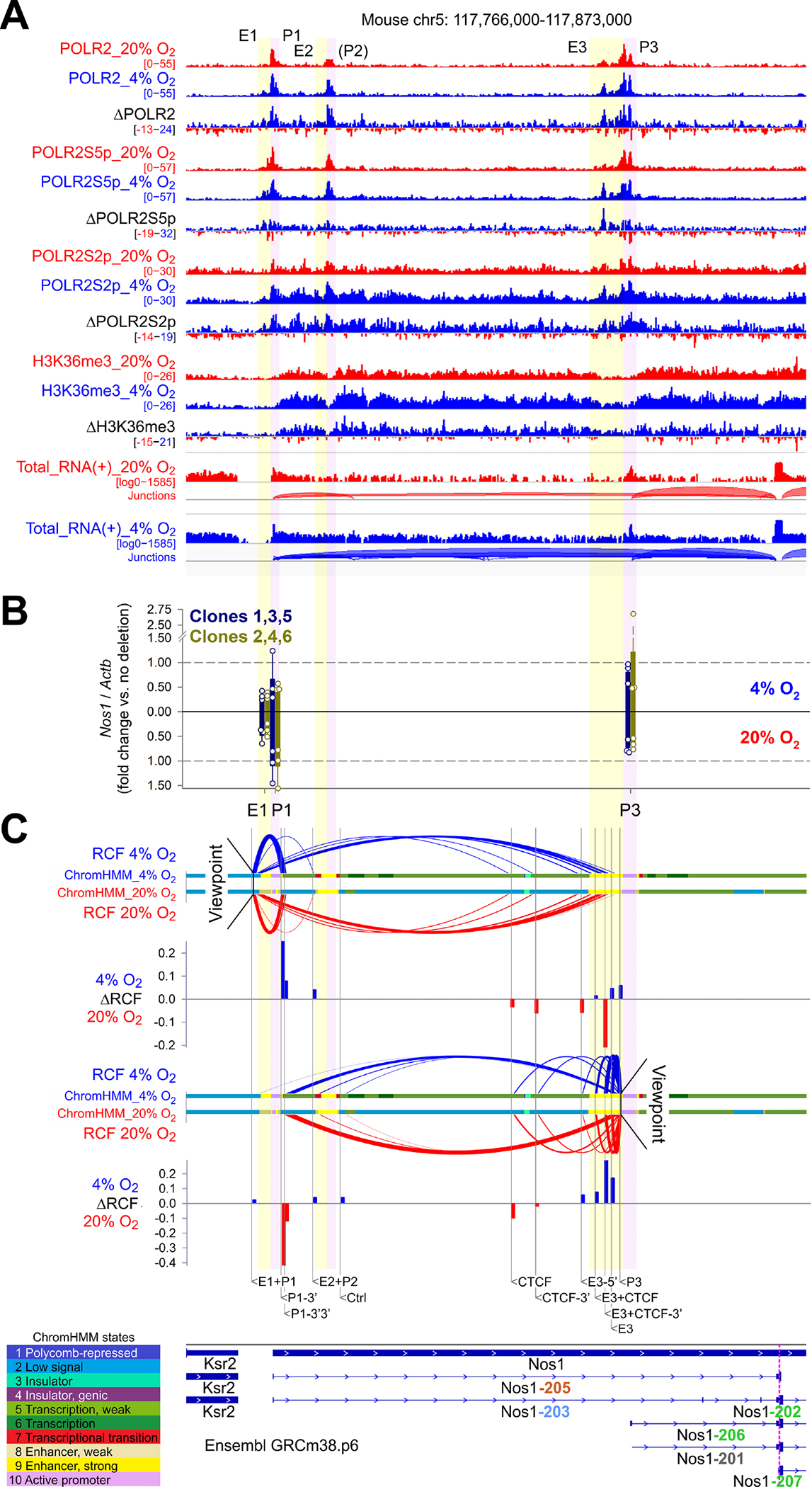
(A) ChIP-seq and total RNA-seq (+ strand) profiles in the Ensembl GRCm38.p6 Nos1 locus of N1E-115 cells cultured at 20% O2 (red) or 4% O2 (blue) for 3 days. Aggregate data from 2 (ChIP-seq) or 3 (RNA-seq) replicates and subtracted (4% O2−20% O2) data are displayed. (B) Nos1 expression in N1E-115 cells bearing CRISPR-Cas9 deletions in E1, P1, and P3 (two clones/position, n=3 cultures/clone). Mean±SD data from cells cultured at 4% (upright) and 20% O2 (inverted) are shown. (C) Enhancer-promoter loops detected by 3C using NcoI. qPCR data from 3 experiments were expressed relative to genomic DNA representing the same locus expressed in bacterial artificial chromosome (BAC). BAC-normalized data were scaled to a range of 0–1 across the loci tested to compare data obtained at 4% O2 (upright) and 20% O2 (inverted). RCF, relative crosslinking frequency (scaled 2ΔCT vs. BAC). The colored ribbons identify epigenetic states discovered by ChromHMM using a 10-state model. Bar plots show ΔRCF values (4% O2−20% O2).
