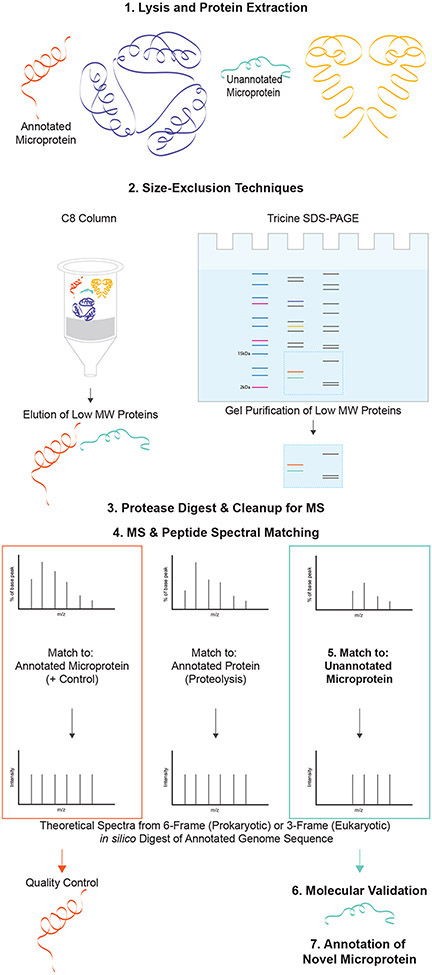
FIGURE 3.
Mass Spectrometry Workflow for Detection of Unannotated Microproteins. To search for novel microproteins in a sample of interest, low molecular weight proteins are isolated from total protein after cell lysis. Size-exclusion techniques include, but are not limited to, solid-phase extraction and polyacrylamide gel electrophoresis techniques. Low molecular weight protein is digested with a protease, producing a sample of uniform peptide length appropriate for mass spectrometric (MS) analysis. Experimental spectra are generated and matched to theoretical spectra from a custom database using proteomics software. Detection of annotated microproteins known to be expressed in the system of interest can serve as a positive control for success of small protein enrichment and known small proteome coverage, but these spectra are otherwise computationally excluded. Peptides deriving from proteolysis of canonical proteins before size-exclusion are computationally identified and excluded from consideration. High scoring experimental spectra without any matches to known microproteins can be subjected to further molecular validation, leading to annotation of novel microproteins.