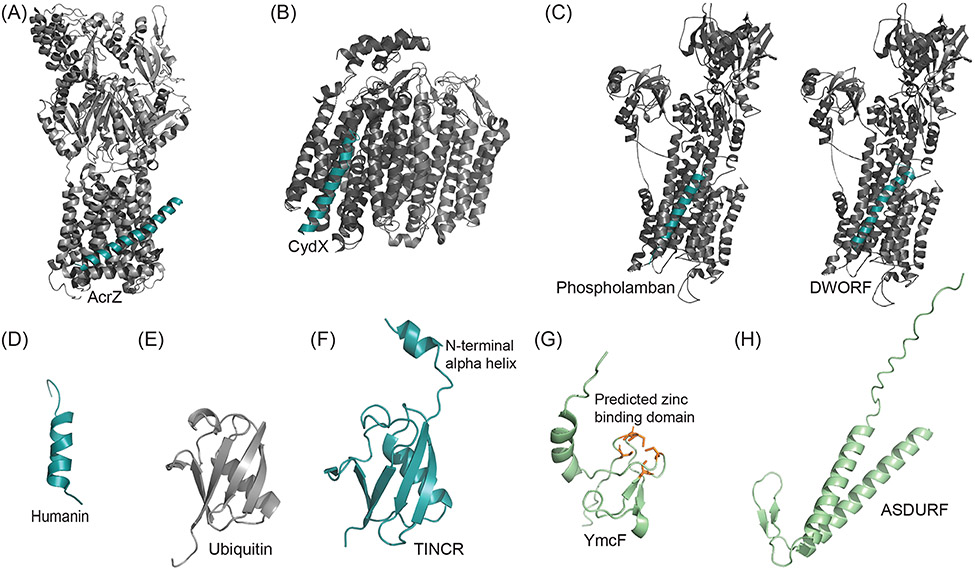
FIGURE 4.
Experimentally Determined Microprotein Structures. (A) Crystal structure of AcrB (grayscale) of the tolC efflux pump in complex with microprotein AcrZ (cyan). PDB: 5NC5. (B) Cryo-EM structure of bacterial microprotein CydX (cyan) in complex with transmembrane cytochrome bd-I oxidase (grayscale). PDB: 6RKO. (C) Crystal structure of SERCA1a calcium pump (grayscale) with bound single-pass transmembrane microprotein phospholamban (cyan), which downregulates SERCA activity. PDB: 4Y3U. Solid-state NMR structure of helix-loop-helix microprotein DWORF (cyan) modeled into SERCA1a calcium pump (grayscale) based on Venkateswara et al. 2022. PDB: 4Y3U, 7MPA. (D) NMR structure of wild-type humanin in 30% 2,2,2-trifluoroethanol (organic) solution. PBD: 1Y32. (E) Crystal structure of Ubiquitin monomer. PDB: 1AAR. (F) Crystal Structure of ubiquitin-like TINCR microprotein with additional N-terminal alpha helix. PDB: 7MRJ. (G) Predicted structure of bacterial microprotein YmcF generated with AlphaFold, obtained from UniProt[166] (green). Five cysteines (orange) in the YmcF sequence are predicted to form a zinc-finger domain common to RNA binding proteins. (H) Predicted structure of PAQosome binding microprotein ASDURF generated with AlphaFold, obtained from UniProt[166].