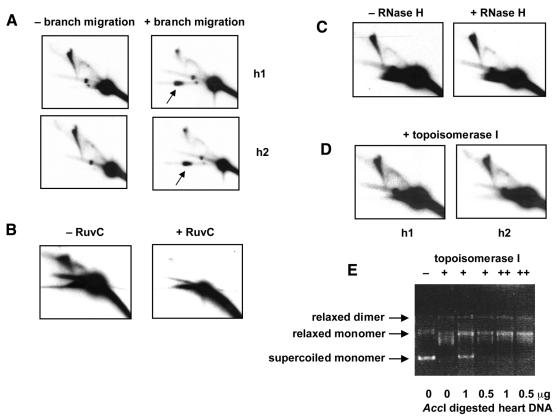
Fig. 3. Resolution of junctional molecules. 2DNAGE of AccI-digested human heart DNA samples (not S1-digested), probed for the 4.8 kb OL-containing fragment, after various treatments. (A) Samples h1 and h2 with or without treatment of the first-dimension gel slab to promote branch migration. Note the large increase in material from the X-spike now migrating in the second dimension with the mobility of the unit fragment (arrow). (B) Sample h2 (2 µg) incubated for 20 h in agarose gel plug with or without RuvC. Note the increase in heterogeneous double-stranded DNA digestion products running below the unit fragment, after RuvC treatment. (C) Sample h1 (2 µg) incubated for 1 h with or without 1 U RNase H. (D) Samples h1 and h2 treated with topoisomerase I prior to electrophoresis. (E) Validation of topoisomerase assay. Supercoiled plasmid DNA (500 ng) incubated either with (lane 1) or without (lane 2) 3 U topoisomerase I, or with 15 U of the enzyme in the presence of AccI-digested heart DNA as shown. Lanes marked ++ were incubated with an additional 6 U of the enzyme for a further 2 h, after overnight reactions. The selected conditions used for the main experiment completely remove supercoiling of the test plasmid, and hence should also separate any catenated molecules.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.