SUMMARY
The dorsal striatum is organized into functional territories defined by corticostriatal inputs onto both direct and indirect spiny projection neurons (SPNs), the major cell types within the striatum. In addition to circuit connectivity, striatal domains are likely defined by the spatially determined transcriptomes of SPNs themselves. To identify cell-type-specific spatiomolecular signatures of direct and indirect SPNs within dorsomedial, dorsolateral, and ventrolateral dorsal striatum, we used RNA profiling in situ hybridization with probes to >98% of protein coding genes. We demonstrate that the molecular identity of SPNs is mediated by hundreds of differentially expressed genes across territories of the striatum, revealing extraordinary heterogeneity in the expression of genes that mediate synaptic function in both direct and indirect SPNs. This deep insight into the complex spatiomolecular organization of the striatum provides a foundation for understanding both normal striatal function and for dissecting region-specific dysfunction in disorders of the striatum.
Graphical Abstract
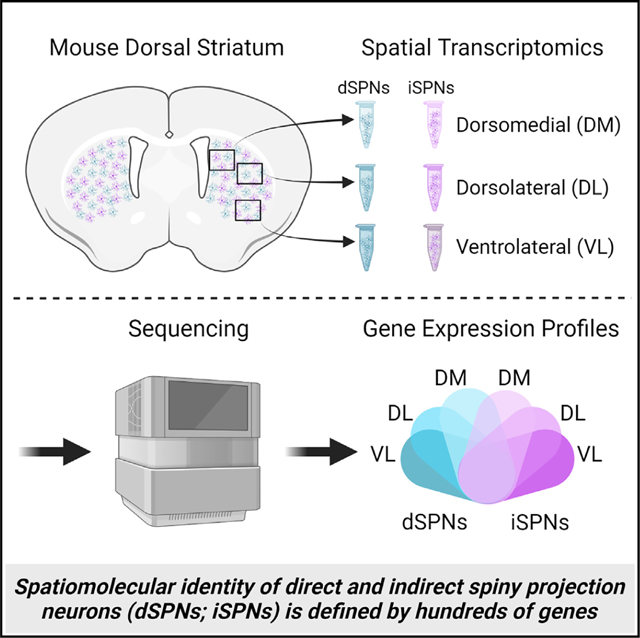
In brief
Roman et al. demonstrate that the molecular identity of spiny projection neurons within the dorsal striatum is mediated by hundreds of genes that are differentially expressed among the dorsomedial, dorsolateral, and ventrolateral striatum, revealing complex spatiomolecular organization in the expression of genes that mediate synaptic function across striatal territories.
INTRODUCTION
The homogeneous appearance of the dorsal striatum at the macroscopic level belies its complex organization across functional, anatomical, cellular, and molecular domains. Because the dorsal striatum receives widespread afferents from the neocortex, it subserves both executive and sensorimotor functions whereby the dorsomedial striatum (analogous to the primate caudate) mediates goal-directed behavior, and the dorsolateral striatum (analogous to the putamen) mediates sensorimotor function. The functional domains are defined, in part, according to the organization of corticostriatal afferents. The dorsomedial or associative striatum receives convergent cortical innervation from association cortices such as anterior cingulate and orbitofrontal areas.1–5 In contrast, the lateral region of the dorsal striatum receives somatotopically organized cortical input predominantly from sensorimotor areas,3–5 so that in rodents, the dorsal portion corresponds to the trunk and lower limbs, whereas the ventral portion corresponds to the upper limbs and orofacial regions. The functional heterogeneity of the dorsal striatum is reflected in the diversity of neurologic, psychiatric, and behavioral disorders associated with striatal dysfunction, including Parkinson’s disease, dystonia, schizophrenia, obsessive compulsive disorder, autism spectrum disorder, and drug addiction.
The striatum is also organized at the cellular level. The majority of striatal neurons are spiny projection neurons (SPNs), which receive glutamatergic innervation from cortico- and thalamostriatal neurons and dopaminergic innervation from nigrostriatal neurons. There are two types of SPNs that are broadly classified by their long-range efferents. In general, direct pathway SPNs (dSPNs) project to internal pallidum/substantia nigra reticulata to promote movement, whereas indirect pathway SPNs (iSPNs) project to the external pallidum to inhibit movement.6–8 Despite their distinct efferent targets and functions, dSPNs and iSPNs are intermixed within the striatum and are morphologically indistinguishable. However, SPN subtypes can be distinguished at the molecular level. dSPNs express a unique subset of genes including Drd1 (D1 dopamine receptors) and Tac1 (protachykinin), while iSPNs express Drd2 (D2 dopamine receptors), Adora2a (A2a adenosine receptors), and Penk (proenkephalin), among others.
In addition to cell-type-specific gene expression, several genes that exhibit differential expression across striatal axes have been identified using single-cell RNA sequencing (scRNA-seq) of dissociated striatal tissue combined with advanced bioinformatic analyses.9–13 The differential levels of expression appear to correlate with the organization of corticostriatal projections,12 suggesting that the functional domains of the striatum may be shaped by both circuit connectivity and the molecular signatures of SPNs. However, considering the complex functional organization of the dorsal striatum, surprisingly few spatially mediated mRNAs have been identified. Therefore, to provide a detailed map of the spatiomolecular signatures of SPNs within the dorsal striatum, we used spatial transcriptomics to identify the cell-type-specific transcriptome of dSPNs and iSPNs from functional territories of the dorsal striatum. Spatial transcriptomics uses multiplexed in situ hybridization for cell-type-specific interrogation of protein coding genes while maintaining the spatial organization of the tissue.14 Using this approach, we identified hundreds of genes in both dSPNs and iSPNs that are differentially expressed across territories of the dorsal striatum, revealing the remarkable heterogeneity in the transcriptomes of SPNs.
RESULTS
Spatial transcriptomics gene expression profiles were consistent with the known transcriptional signatures of dSPNs and iSPNs
The intermediate striatum (Figure 1A) was selected for analysis because it receives projections from all cortical areas and exhibits subregions that are delineated by both cortical innervation and behavioral function.4 As such, samples were collected from three striatal subregions, including the associative or dorsomedial, and two regions of the sensorimotor striatum: dorsolateral, which corresponds to the trunk and limbs, and ventrolateral, which corresponds to the upper limbs and orofacial regions (Figure 1B). Fluorescent in situ hybridization for D1 dopamine receptor (Drd1) and A2a adenosine receptor (Adora2a) mRNA was used to label dSPNs and iSPNs, respectively, for cell-type-specific sample collection (Figures 1B and 1C). A total of 11,477 transcripts were detected across all samples. The majority of cells sampled were neuronal with little representation of non-neuronal cells such as glia (Figure S1). The total number of genes detected among striatal subregions differed, with the greatest number of genes detected in the ventrolateral striatum in both dSPNs and iSPNs (Figure 1D). The density of dSPNs and iSPNs was not significantly different among striatal regions (one-way repeated measures ANOVAs: dSPNs: F1.201, 6.005 = 3.483, p = 0.11; iSPNs: F1.324, 6.622 = 2.595, p = 0.15). Further, the number of genes detected within each striatal region was comparable between dSPNs and iSPNs.
Figure 1. Spatial transcriptomics gene expression profiles were consistent with the known transcriptional signatures of dSPNs and iSPNs.
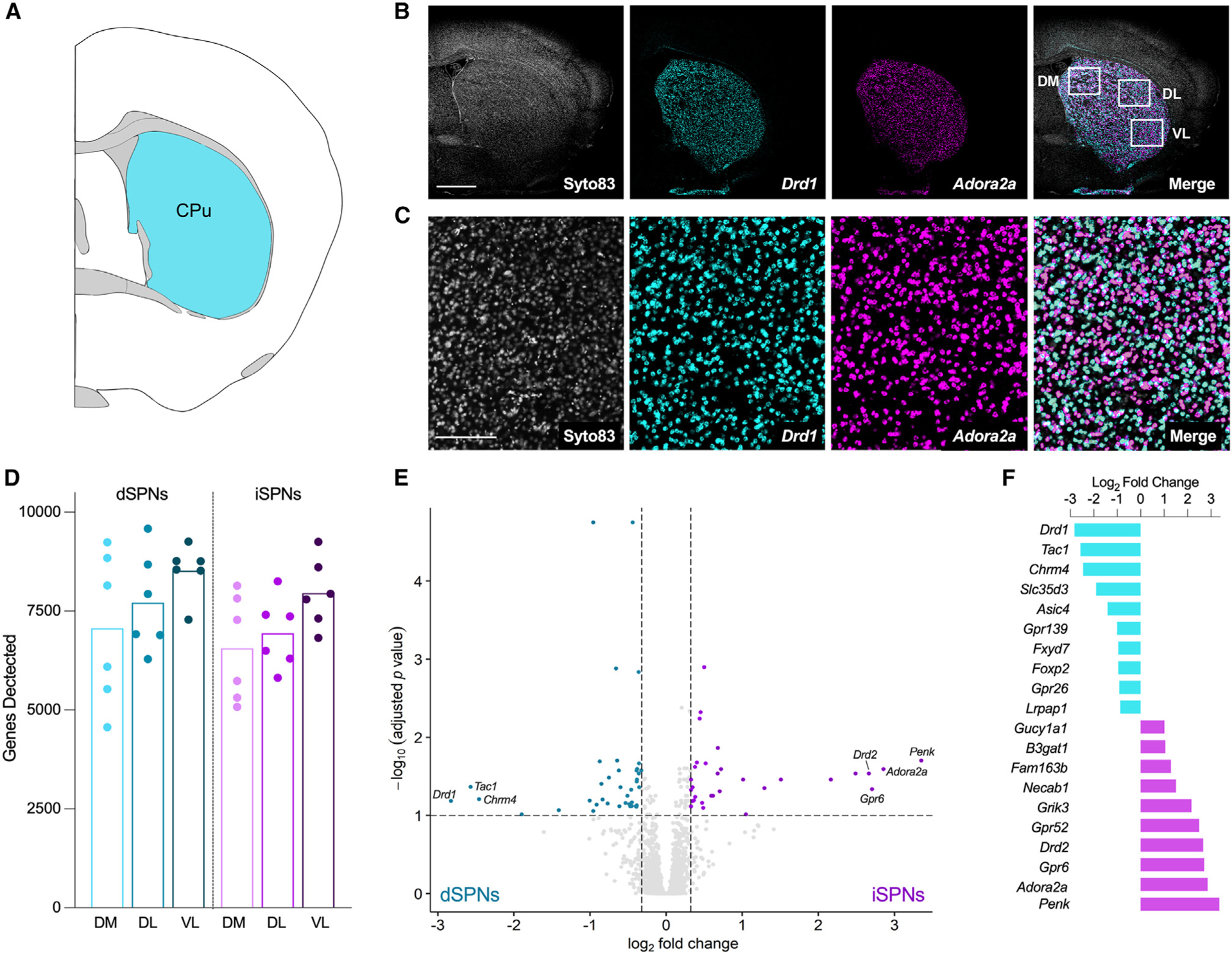
(A) Schematic coronal section adapted from the Allen Brain Reference Atlas (image 53; 0.14 mm from bregma) depicting the striatal sampling region.
(B) Representative micrographs depicting the coronal section of striatum used for sample collection. Nuclei were visualized with Syto83 (gray), dSPNs were identified by fluorescent in situ hybridization for the D1 dopamine receptor (Drd1; aqua), and iSPNs were identified by fluorescent in situ hybridization for the adenosine A2a receptor (Adora2a; purple). White boxes in the merged micrograph demarcate the dorsomedial (DM), dorsolateral (DL), and ventrolateral (VL) striatal subregions used for sample collection. Scale bar represents 1 mm.
(C) Higher power images of the sections shown in (B). Scale bar represents 200 μm.
(D) Number of genes detected in each sample (n = 6, three striatal subregions/mouse). Each point represents an individual mouse, and bars represent means. There was an effect of striatal subregion, with the greatest number of genes detected in the ventrolateral striatum, but the number of genes detected was comparable between dSPNs and iSPNs (two-way RM ANOVA, main effect of region, F2,20 = 12.7, p = 0.0003; main effect of cell type, F1, 10 = 0.9641, p = 0.3493).
(E) Volcano plot depicting genes differentially expressed in dSPNs and iSPNs collapsed across regions. Genes enriched in dSPNs (FC < −1.25, n = 39) are indicated by aqua dots, and genes enriched in iSPNs (FC > 1.25, n = 31) are indicated by purple dots (unpaired t tests with Benjamini-Hochberg correction for multiple comparisons p-adj <0.1). All other genes are represented by light gray dots.
(F) The 10 most-enriched genes in dSPNs (aqua) and iSPNs (purple), based on log2 fold change.
To determine if the spatial profiling had enriched for mRNAs from each SPN subtype, we examined the transcriptional signatures of the dSPN and iSPN samples. Seventy genes were differentially expressed between dSPNs and iSPNs, which is in line with a previous study in mice.9 The expression of 39 genes was enriched in dSPNs, including the well-known dSPN markers Drd1, Tac1, and Chrm4. The expression of 31 genes was enriched in iSPN samples, including the well-known iSPN markers Penk, Adora2a, Gpr6, and Drd2 (Figures 1E and 1F). These results demonstrate that the digital spatial profiling effectively captured and enriched mRNA probes from either dSPNs or iSPNs.
Differential gene expression across dorsal striatal subregions in dSPNs and iSPNs
We first examined the spatial patterns of gene expression in dSPNs. Across the medio-lateral axis of the striatum, 45 genes were differentially expressed between the dorsomedial and dorsolateral striatum with 15 genes enriched in the dorsomedial region and 30 genes enriched in the dorsolateral region (Figure 2A; fold change (FC) ≥ 1.5 and p-adj < 0.05). In agreement with studies in mice and marmosets,10,11 Col6a1 and Crym were highly enriched in the dorsomedial striatum, while Cnr1 and Gpr155 were highly enriched in the dorsolateral striatum, providing additional validation for the approach. We next examined differences in dSPN gene expression across the dorsoventral axis by comparing dorsolateral to ventrolateral striatum. In total, 37 genes were differentially expressed with 22 genes enriched in the dorsolateral striatum and 15 genes enriched in the ventrolateral striatum (Figure 2C). A comparison of dSPN gene expression between dorsomedial and ventrolateral striatum, which are anatomically distant subregions, revealed the extent of the regional heterogeneity in the transcriptome within the dorsal striatum, whereby 399 genes were differentially expressed between dorsomedial and ventrolateral striatum: 219 genes were enriched in the dorsomedial striatum, and 180 were enriched in ventrolateral striatum (Figure 2E).
Figure 2. Differential gene expression across dorsal striatal subregions in dSPNs and iSPNs.
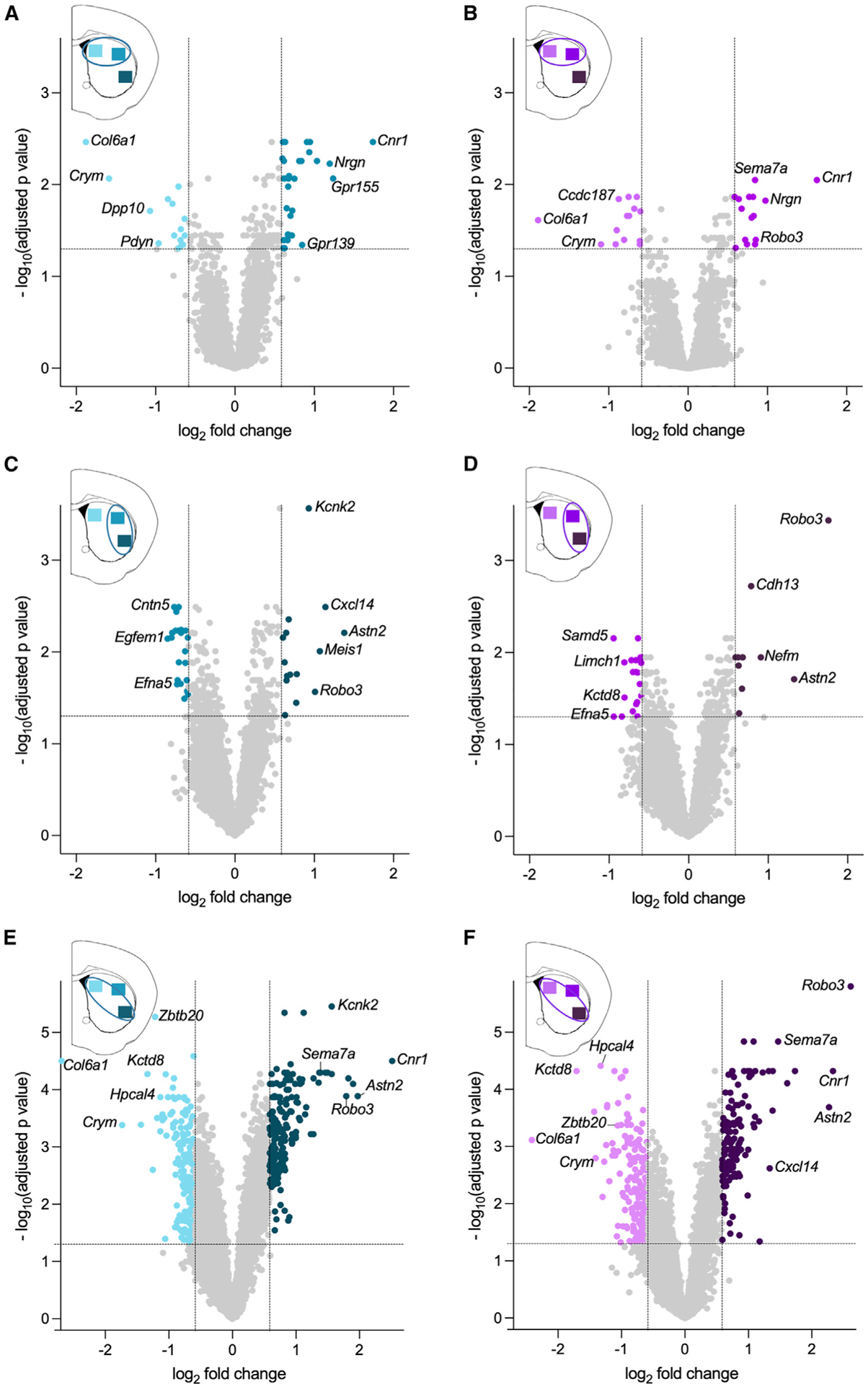
(A) dSPN genes enriched in dorsomedial striatum (light aqua) versus dorsolateral striatum (medium aqua).
(B) iSPN genes enriched in dorsomedial striatum (light purple) versus dorsolateral striatum (medium purple).
(C) dSPN genes enriched in dorsolateral striatum (medium aqua) versus ventrolateral striatum (dark aqua).
(D) iSPN genes enriched in dorsolateral striatum (medium purple) versus ventrolateral striatum (dark purple).
(E) dSPN genes enriched in dorsomedial striatum (light aqua) versus ventrolateral striatum (dark aqua).
(F) iSPN genes enriched in dorsomedial striatum (light purple) versus ventrolateral striatum (dark purple). Aqua or purple dots denote genes enriched between striatal regions (FC > 1.5 indicated by vertical lines, and p-adj <0.05 indicated by the horizontal line). All other genes are represented by light gray dots. Data are from six mice with three regions sampled/mouse.
A similar pattern was observed when gene expression in iSPNs was compared across striatal regions. Twenty-nine genes were differentially expressed between the dorsomedial and dorsolateral striatum with 14 genes enriched in the dorsomedial, and 15 genes were enriched in dorsolateral (Figure 2B). In agreement with the dSPN data, Col6a1 and Crym, which are genes known to be expressed in both dSPNs and iSPNs, were highly enriched in dorsomedial striatal iSPNs. Likewise, Cnr1 was enriched in dorsolateral striatum. In the dorsoventral axis, 31 genes were differentially expressed between dorsolateral and ventrolateral striatum; 20 genes were dorsolateral enriched, and 11 genes were ventrolateral enriched (Figure 2D). Finally, a comparison between dorsomedial and ventrolateral iSPNs revealed 350 genes that were differentially expressed, with 208 genes enriched in dorsomedial striatum and 142 genes enriched in ventrolateral striatum (Figure 2F).
Patterns of gene expression across striatal regions
To identify broad patterns of expression across the dorsal striatum, we focused on genes that exhibited FC >2 in expression (p-adj < 0.05) among the striatal subregions, including 44 genes from the dSPN dataset and 42 genes from the iSPN dataset (Figures 3A–3D and S2; Table S1). Within this subset were 10 genes previously identified as molecular markers of intrastriatal spatial organization, including Cnr1, Coch, Col6a, Cpne6, Crym, Cxcl14, Gpr139, Gpr155, Nefm, and Syt10,10,12 while the remaining ~75% had not been previously recognized as spatially mediated. Dorsomedial-ventrolateral differential expression occurred among all genes with FC >2.0 across striatal territories (Figures 3A–3D) with DM > DL > VL for some genes (Figures 3A and 3C) and VL > DL > DM for others (Figures 3B and 3D). Although the differential expression was not always a linear gradient across regions, we did not observe genes for which expression in DL was higher than both DM and VL, even when the criterion was expanded to include genes with FC >1.5 across striatal territories. To validate the spatial distribution of mRNA expression, we examined the anatomical distribution of mRNA expression within the striatum using independently performed in situ hybridization from the Allen Brain Atlas (Figure 3E). In situ hybridization corroborated the results of the RNA spatial profiling, illustrating differential expression of both dorsomedial to ventrolateral and ventrolateral to dorsomedial mRNA expression.
Figure 3. Patterns of gene expression across striatal regions.
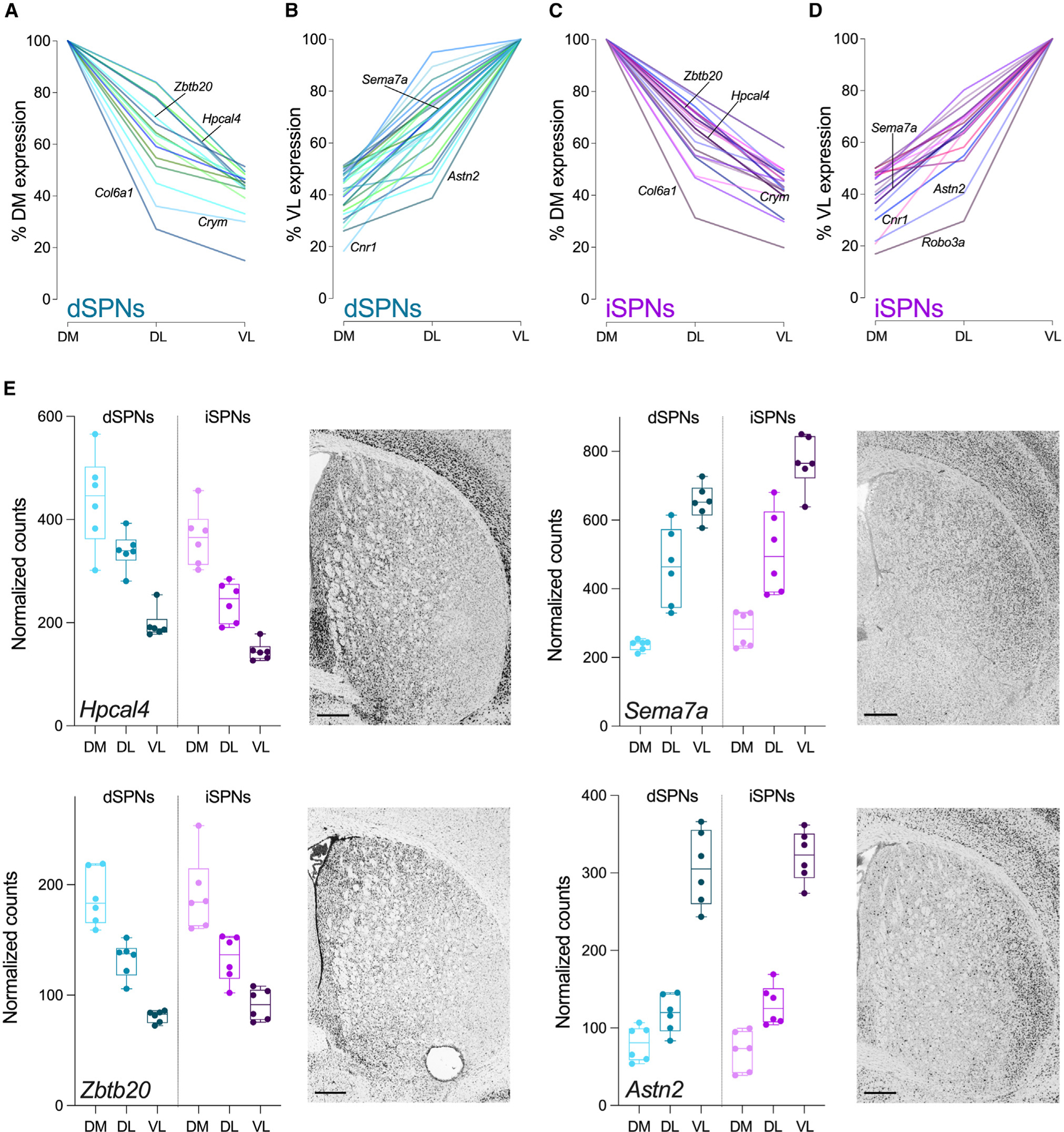
(A‒D) Summary plots of the expression profiles for all genes with FC >2 across striatal subregions. (A) Genes with DM > VL and (B) genes with VL > DM expression levels in dSPNs. (C) Genes with DM > VL and (D) genes with VL > DM expression levels in iSPNs. Gene labels correspond to genes in (E) or genes previously identified as molecular markers of intrastriatal organization. Data are expressed as a percent of DM (A and C) or VL (B and D) expression for each gene.
(E) Representative genes with spatial patterns of expression in dSPNs and iSPNs. All genes have FC >2 (linear mixed models with Benjamini-Hochberg p-adj <0.05) in expression between dorsomedial and ventrolateral striatum in dSPNs and iSPNs. Adjacent in situ hybridization images are at the same anterior-posterior level as the brain sections used for spatial transcriptomics (http://mouse.brain-map.org). Scale bars represent 400 μm. Box and whiskers plots with individual datapoints depict the median (horizontal line) with the box extending from the 25th to the 75th percentiles and whiskers extending to maximum and minimum for data obtained from six mice with three striatal regions sampled/mouse (DM, dorsomedial; DL, dorsolateral; VL, ventrolateral).
It has been suggested that the spatial organization of mRNA expression within the striatum defines the striatal subregion regardless of SPN subtype.10 For example, genes enriched in dorsomedial striatum compared to ventrolateral striatum would be expected to be enriched in both dSPNs and iSPNs. This hypothesis was based on a relatively limited number of regionally expressed genes identified using scRNA-seq in dissociated striatal cells. That our RNA spatial profiling assay identified hundreds of differentially expressed genes among striatal subregions while distinguishing between dSPNs and iSPNs in each region provided the opportunity to test this hypothesis. Therefore, we asked if genes that were differentially expressed by >1.5-fold between dorsomedial and ventrolateral striatum (399 for dSPNs and 350 for iSPNs) exhibited similar patterns of expression in both dSPNs and iSPNs. The vast majority of differentially expressed genes were regulated in parallel across the striatum in dSPNs and iSPNs. Indeed, only 24 genes did not appear to follow this pattern whereby region-dependent expression was observed in either dSPNs or iSPNs (Table S2).
Region-specific neuronal features of SPNs
Because the spatial transcriptomics provided a rich dataset consisting of thousands of genes within identified striatal subregions, it was possible to ascribe biological context to the regional heterogeneity in SPN mRNA expression. To provide insight into the potential functional ramifications of the hundreds of differentially expressed genes across the striatum, regional variation in neuronal characteristics was identified using Gene Ontology (GO) analyses comparing dorsomedial- to ventrolateral-enriched genes. For both dSPNs and iSPNs, the “cellular component” terms, which describe the subcellular location of the gene products, included the distinguishing features of a neuron, particularly SPNs, such as “dendritic spine” and ‘GABAergic synapse.” Terms describing the fundamental building blocks of a cell such as nucleus, endoplasmic reticulum, or mitochondria were absent, suggesting that the regional differences in SPNs are driven by features that define neuronal function per se. Indeed, the terms were largely focused on the synapse for both dSPNs and iSPNs (Figures 4A and 4B). An analysis of the genes associated with “intrinsic component of presynaptic membrane,” the top cellular pathway for both dSPNs and iSPNs (Figure 4C), revealed differential expression across striatal subregions for genes implicated in neurotransmission including receptors (Adora2a, Chrm4, Chrna4, Cnr1, Drd2, Grin1, Grin2a, Gria3, Grm3, Htr2a, Oprk1, P2ry1), cell-cell interactions (Adam23, Adgrl1, Cadm1, Cdh9, Cntn5, Cntnap4, Nptn, Pcdh17), and calcium homeostasis (Atp2b4, Atp2b2, Atp2b1). The dorsomedial-ventrolateral differential expression of genes associated with neuronal signaling was consistent with the “molecular function” GO analysis, where the terms were predominantly focused on cation channel activity (Figures 4D and 4E). The majority of the genes associated with the term “voltage-gated cation channel activity,” which was identified for both dSPNs and iSPNs, were potassium channels or potassium channel modifiers (Kcna5, Kcnab1, Kcnb2, Kcnd3, Kcnf1, Kcng1, Kcnh5, Kcnip2, Kcnj6 Kcnk1, Kcnk2, Kcnq3, Kcnt1); several calcium channel subunits (Cacna1c, Cacna1i, Cacna2d3, Cacnb2) were also included in this term (Figure 4F).
Figure 4. Region-specific neuronal features of SPNs.
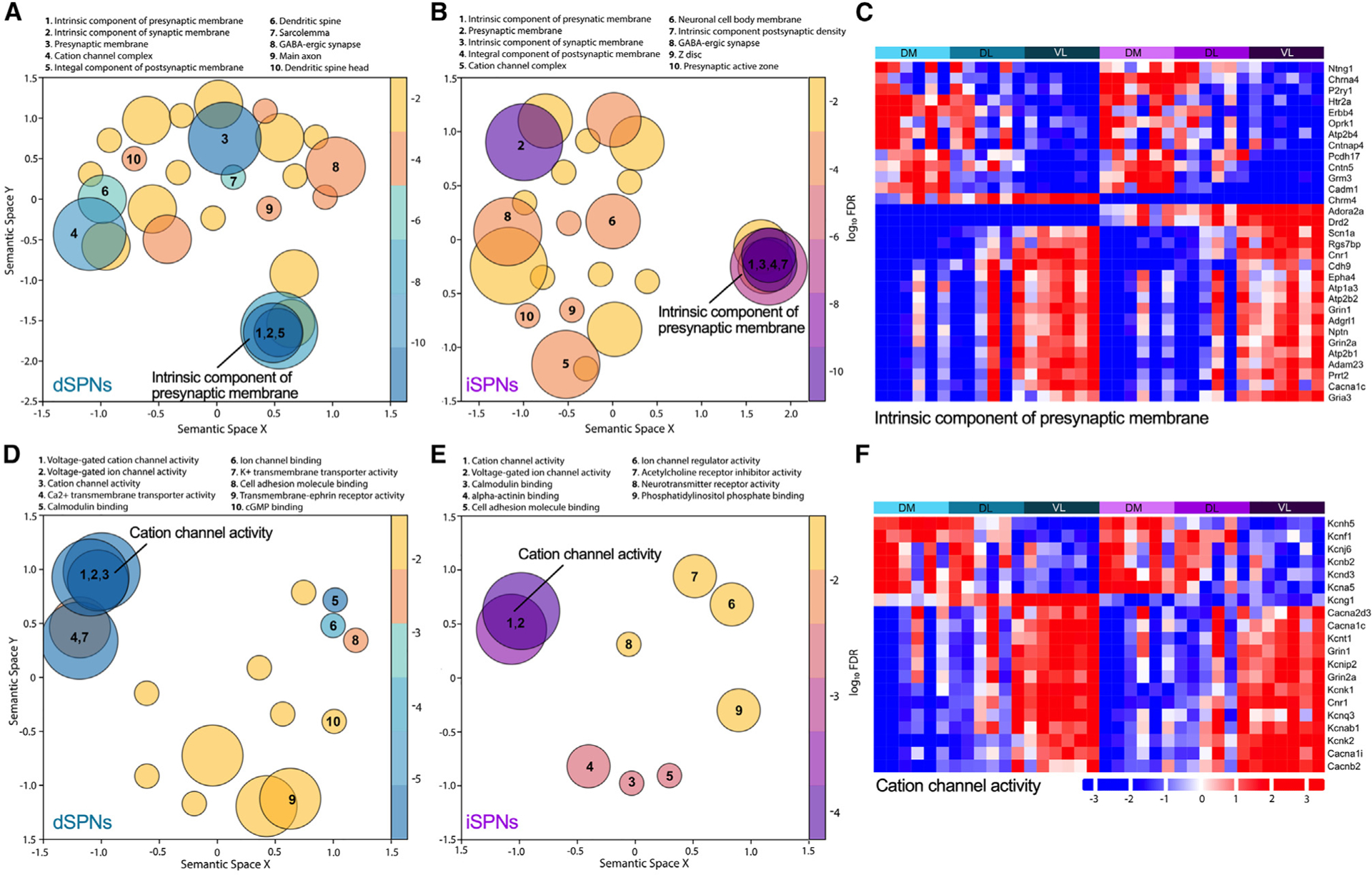
(A, B, D, and E) Gene Ontology (GO) analysis of dorsomedial versus ventrolateral striatum for dSPNs and iSPNs. Cellular component terms for dSPNs (A) and iSPNs (B) and molecular function terms dSPNs (D) and iSPNs (E). Terms are represented by circles scaled to the number of genes represented within the term. Overlapping circles indicate genes shared within the terms. False discovery rate adjusted p values from the GO enrichment analysis are represented using a gradient color palette. The top 10 pathways are numbered.
(C and F) Heatmaps illustrate the regional distribution of genes identified in the GO terms “intrinsic component of presynaptic membrane” (C) and “cation channel activity” (F). The colors from blue to red in the heatmap indicate the increasing expression of genes. Bars above the heatmap denote region and cell type with aqua for dSPNs and purple for iSPNs and abbreviations for dorsomedial (DM), dorsolateral (DL), and ventrolateral (VL) striatal subregions. Each column within a region represents an individual mouse. Only genes with FC ≥1.5 between groups (dorsomedial vs. dorsolateral, dorsomedial vs. ventrolateral, or dorsolateral vs. ventrolateral) are included in the heatmaps.
DISCUSSION
Our results provide deep insight into the highly complex spatiomolecular organization of striatal dSPNs and iSPNs, revealing that the molecular identity of SPNs is mediated by hundreds of differentially expressed genes across the medial-lateral axis of the striatum. Although recent advances in cell-type-specific RNA sequencing have suggested that transcriptional heterogeneity correlates with SPN position within the striatum,9–13 the results presented here demonstrate the extent of the spatially mediated heterogeneity. Of the >60 genes with >2-fold difference in expression between dorsomedial and ventrolateral striatum, only 10 were previously known to exhibit differential expression across the dorsal striatum.10,12 When considering genes with >1.5-fold difference in expression between dorsomedial and ventrolateral striatum, hundreds were identified, revealing the extraordinary scope of the regional variation in gene expression within the dorsal striatum. More than 90% of differentially expressed genes in dSPNs and iSPNs were mediated in parallel across the medial-lateral axis of the dorsal striatum in support of the hypothesis that the molecular signature of an SPN conveys striatal coordinates.10 This rich dataset also revealed the broader biological implications of mRNA expression across striatal territories, providing a unique resource for dissecting the region-specific cellular, physiologic, and behavioral properties of the dorsal striatum. To that end, we provide the entire normalized dataset in a format that is easily mined and analyzed (Data S1).
There were significant regional differences in the expression of genes that mediate both synaptic function and ion channels, suggesting that the physiological and signaling properties of SPNs vary across striatal territories. Indeed, fluctuations in membrane potentials differ between dorsomedial and dorsolateral SPNs. SPNs shift between two states: a depolarized “up state” near the action potential threshold and a more polarized “down state,” which are thought to contribute to the selection and integration of corticostriatal inputs.15 The up state membrane potential in dorsomedial SPNs is higher than dorsolateral SPNs.16 The membrane potential attained during the up state is governed by voltage-gated potassium channels and is dependent on the activation of R- or T-type calcium channels and NMDA receptors.17,18 That the GO term “voltage-gated ion channel activity” identified multiple voltage-gated potassium channels, T-type calcium channels (Cacna1i), and NMDA receptors (Grin1 and Grin2a) with differential expression across striatal territories provides a plausible explanation for the difference in up-state membrane potentials between regions.
Numerous neurotransmitter receptors were differentially expressed across striatal regions. Notably, many of these receptors mediate activity-dependent changes in the strength of corticostriatal synapses, including type 1 cannabinoid receptors (Cnr1), D2 dopamine receptors (Drd2), M4 muscarinic receptors (Chrm4), and NMDA receptors (Grin1, Grin2a).19–22 Synaptic plasticity, such as corticostriatal long-term depression (LTD), varies according to striatal subregion. LTD in the dorsomedial striatum is NMDA receptor dependent and does not require either type 1 cannabinoid receptors or D2 dopamine receptor signaling.23 However, LTD in the dorsolateral striatum depends on the activation of type 1 cannabinoid receptors and D2 dopamine receptors,24,25 consistent with the high expression of type 1 cannabinoid receptors and D2 dopamine receptors in the dorsolateral striatum compared to dorsomedial striatum. Although studies that directly compare the physiological properties of dorsomedial to dorsolateral SPNs are limited, the spatiomolecular results presented here in combination with the known properties of dorsomedial vs. dorsolateral physiology suggest that the identification of region-specific physiological signatures will be critical for understanding signal processing within striatal territories.
Disorders associated with striatal dysfunction often exhibit subregion-selective abnormalities. Dorsal to ventral gradients in connectivity are observed in psychosis with a reduction in functional connectivity between the dorsal caudate and prefrontal cortex and an increase between the ventral caudate and orbitofrontal cortex.26 Notably, we identified significant territorial differences in the expression of genes associated with rare variants in schizophrenia, including Grin2a (NMDA receptor type 2A subunit) and Gria3 (AMPA receptor type 3 subunit).27,28 Gradients of dysfunction are also observed in Huntington’s disease where the striatal neuropathology and neurodegeneration first appear in the medial caudate nucleus, the tail of the caudate, and dorsal putamen and progressively affect more lateral and ventral regions of the striatum.29 Other disorders associated with the striatal dysfunction are characterized by neuroadaptations within specific striatal territories. In a model of dystonia, dopamine receptor-mediated intracellular signaling is hyper-responsive in the dorsomedial, whereas the dorsolateral striatum, which receives sensorimotor corticostriatal afferents, is unresponsive, suggesting that region-specific dysfunction contributes to the distinct features of this movement disorder.30 The dorsolateral striatum is specifically implicated in compulsive consumption and drug seeking including cocaine, morphine, nicotine, and alcohol.31–34 Indeed, in individuals with alcohol use disorder and in rats trained to self-administer alcohol, expression of the M4 muscarinic receptor (Chrm4) is downregulated in the putamen (dorsolateral striatum), where Chrm4 is abundantly expressed (Figure 4C), but unaffected in the caudate (dorsomedial striatum). Direct administration of an M4 muscarinic receptor-positive allosteric modulator into the rat dorsolateral striatum to counteract the signaling deficit reduces alcohol self-administration and alcohol seeking.35 Thus, knowledge of the molecular mechanisms underlying striatal organization is not only critical for understanding pathogenesis and pathophysiology, it provides a foundation for the rational design of region-biased therapeutics.
Limitations of the study
The advantage of spatial transcriptomics is that cell-type-specific and region-specific information is preserved, unlike bulk cell-type-specific enrichment processes. However, it does not provide the granularity of scRNA-seq. Laser capture microdissection combined with scRNA-seq could provide both spatial and cellular resolution; whereas the spatial transcriptomics approach presented here collects the sample from identified cell types, microdissection with scRNA-seq requires post hoc identification of cells based on mRNA expression patterns. Further, our analyses were confined to the medial-lateral axis of the intermediate dorsal striatum, which was interrogated because the functional subregions are well defined, and this is a commonly studied region of the dorsal striatum.4 As SPN transcriptomes exhibit some variation along both the rostral-caudal axis and dorsal-ventral axis (dorsal striatum vs. nucleus accumbens), medial-lateral patterns of expression may vary based on specific coordinates within the striatum.36,37 Additionally, although we did not observe sex-specific effects, it is not possible rule out an effect as this study was not powered for sex as a biological variable. Further, the limitation of all transcriptomic analyses is that mRNA expression does not always reflect protein expression.38 Indeed, regional intrastriatal protein expression has not been intensively examined largely due to the lack of reagents such as antibodies or in situ assays, so the GO analyses will require further exploration at the physiologic level. However, in situ receptor binding or enzyme activity assays for type 1 cannabinoid receptors (Cnr1), guanine deaminase (Gda), and angiotensin 1 converting enzyme (Ace), which all exhibit >2-fold differences in regional mRNA expression, demonstrate that striatal protein activity reflects the mRNA distribution identified in this study.39–41
Having established spatial transcriptomics as a viable approach for cell-type- and region-specific transcriptomics within the dorsal striatum, it is now feasible to examine additional features of striatal organization and cell types including region-specific characteristics of striosome/matrix compartments and striatal interneurons, which are potent mediators of SPN activity. Further, the identification of hundreds of genes with differential expression across striatal territories provides a foundation for deciphering the mechanisms that distinguish functional domains in health and disease.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ellen J. Hess (ejhess@emory.edu).
Materials availability
There are no newly generated materials associated with this paper.
Data and code availability
Unprocessed (raw) Digital Spatial Profiling data are publicly available as of the date of publication. The DOI is listed in the key resources table. The entire normalized dataset is available in the supplemental material.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Tyramide (TSA) linked Opal620 | PerkinElmer | FP1495001KT |
| Cyanine 5 dyes | PerkinElmer | NEL745001KT |
| Neutral Buffer Formalin | EMS Diasum | 15740-04 |
| Syto 83 | ThermoFisher | S11364 |
| Critical commercial assays | ||
| Mm-Adora2a-C2 | ACD | 409438-C2 |
| Mm-Drd1-C3 | ACD | 461908-C3 |
| Mouse Whole Transcriptome Atlas RNA detection probes | Nanostring | GMX-RNA-NGSMsWTA-4 |
| Buffer R | Nanostring | N/A |
| Buffer W | Nanostring | N/A |
| GeoMx Seq Code kit | Nanostring | GMX-NGS-SEQ |
| AmPure XP beads | Beckman Coulter | A63881 |
| Multiplex 5X PCR Master Mix | Nanostring | N/A |
| Deposited data | ||
| Unprocessed (raw) data | This paper | https://doi.org/10.15139/S3/HS5BHE |
| Normalized data | This paper; supplement | N/A |
| In situ hybridization images | Allen Brain Atlas | https://mouse.brain-map.org/static/atlas |
| Experimental models: Organisms/strains | ||
| Mus Musculus C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mus Musculus DBA/2J | The Jackson Laboratory | RRID:IMSR_JAX:000671 |
| Software and algorithms | ||
| ShinyGO v0.76 Web App | (Ge et al.)42 | http://bioinformatics.sdstate.edu/go/ |
| ComplexHeatmap v2.14.0 R package | (Gu et al.)43 | https://doi.org/10.18129/B9.bioc.ComplexHeatmap |
| GO-Figure! Python scripts | (Reijnders and Waterhouse)44 | https://gitlab.com/evogenlab/GO-Figure |
| GraphPad Prism v9.4.1 | GraphPad Software, LLC | https://www.graphpad.com/ |
| GeoMx Next Generation Sequencing Pipeline | Nanostring | N/A |
| GeoMx Digital Spatial Profiler Data Analysis Suite | Nanostring | N/A |
| SpatialDecon v1.8.0 R package | Nanostring | https://github.com/Nanostring-Biostats |
| Other | ||
| Bond RX Fully Automated Research Stainer | Leica Biosystems | N/A |
| Hyb EZ II hybridization oven | ACD | 321710/321720 |
| HybriSlip hybridization covers | Grace Bio-Labs | 714022 |
| Illumina NovaSeq | Novogene | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Experimental procedures were approved by the Emory Animal Care and Use Committee and followed the Guide for the Care and Use of Laboratory Animals. Mice were bred at Emory University. Male and female mice (3 males, 3 females; 18 weeks of age) were F1 hybrids of C57BL/6J x DBA/2J, a background strain that we use routinely to maintain genetic models of movement disorders that lack vigor.45,46 Mice were maintained on a 12h light/dark cycle and allowed ad libitum access to food and water. Principal Component Analysis did not reveal sex-specific effects and we did not further investigate the effect of sex.
METHOD DETAILS
mRNA spatial profiling
Digital Spatial Profiling was performed as described.14 The Mouse Whole Transcriptome Atlas was used to bind mRNA targets (NanoString Technologies, Seattle, WA, USA). The Mouse Whole Transcriptome Atlas consists of 20,175 probes to >98% of protein coding genes of the mouse transcriptome with a small number of very abundantly expressed genes removed to provide better detection of rare transcripts.
Sample preparation
Fresh-frozen striatal tissue sections (10 μm) were mounted on Superfrost Plus slides (Fisher Scientific) and stored at −80°C until use. Sections from the intermediate striatum at +0.14 mm from bregma (n = 6 mice) were used for RNA profiling because it receives projections from all cortical areas and has unique functional subregions.4 Prior to experiments, samples were removed from −80°C and immediately dried using a hair dryer on cold setting to remove moisture. Samples were further dried at 37°C for 1 h in a hybridization chamber, then fixed in 4% paraformaldehyde in 0.1M phosphate-buffer saline (PBS) overnight at room temperature. Slides were washed 3 times in PBS prior to loading into the Bond RX Fully Automated Research Stainer (Leica Biosystems) for automated fluorescent in situ hybridization per manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA, USA). To identify dSPNs and iSPNs for subsequent sample collection, the following probes were used: Mm-Adora2a-C2 (ACD, #409438-C2) and Mm-Drd1-C3 (ACD, #461908-C3). Signal was detected with Tyramide (TSA) linked Opal620 (#FP1495001KT, 1:3000) and Cyanine 5 dyes (#NEL745001KT, 1:4500, PerkinElmer, Waltham, MA, USA). Slides were removed from the Leica, rinsed in deionized water, incubated in 10% Neutral Buffer Formalin (NBF) for 5 min, washed twice in 0.1M Tris-Glycine NBF stop buffer, and then rinsed in PBS, all at RT.
Hybridization with RNA detection probes
Tissue sections were then incubated overnight at 37°C with Mouse Whole Transcriptome Atlas RNA detection probes in Buffer R (NanoString Technologies) using a Hyb EZ II hybridization oven (ACD). The DNA oligonucleotide probes were composed of a sequence complementary to a target mRNA joined to a sequencing oligonucleotide via an ultraviolet photocleavable linker. Each unique sequencing oligonucleotide consisted of a gene-specific barcode, Unique Molecular Index (UMI) and primer binding sites. During incubation, slides were covered with HybriSlip Hybridization Covers (Grace BioLabs, 714022). Following incubation, HybriSlip covers were gently removed, and slides were washed in 50% formamide and 2X saline sodium citrate (SSC) for 25 min at 37°C, twice. Tissues were then washed for 5 min in 2X SSC, subsequently blocked in Buffer W (Nanostring Technologies) for 30 min at room temperature in a humidity chamber, and then incubated in 500nM Syto83 (ThermoFisher) for visualization of nuclei. Next, slides were washed twice in fresh 2X SSC then loaded on the GeoMx Digital Spatial Profiler.
Imaging
Entire slides were imaged, and 3 regions of interest (660 × 785 μm) were identified for each subject in the dorsomedial, dorsolateral, and ventrolateral regions of the striatum. Two areas of illumination (AOIs) were defined within each region: the first AOI contained fluorescent signal for Drd1a and the second contained fluorescent signal for Adora2a. The number of nuclei in each AOI was estimated using the GeoMx software, which counts distinct elements in the DAPI channel; >350 cells were sampled/AOI. After AOIs were defined, the Digital Spatial Profiler exposed each AOI to 385 nm light to release the indexing oligonucleotides. To achieve separation between the transcripts from Drd1a- and Adora2a-expressing cells, the cell-types were exposed to the UV light sequentially by applying a mask to allow the UV light to specifically target one cell-type and then the other, based on the fluorescent profile. After UV exposure, the released indexing oligonucleotides from the entire AOI were collected with a microcapillary. Thus, samples for both dSPNs and iSPNs in all three striatal regions of interest were collected from the same tissue section. Indexing oligonucleotides were deposited in a 96-well plate, dried down overnight and resuspended in 10 μL of DEPC-treated water.
Sequencing
Sequencing libraries were generated by PCR using the indexing oligonucleotides photo-released from the Mouse Whole Transcriptome Atlas RNA detection probes for each AOI. The PCR primer mix (GeoMx Seq Code kit, Nanostring) consisted of primer pairs complementary to the photo-released indexing oligonucleotide that are ligated to unique i5 and i7 index sequences, allowing libraries to be pooled (multiplexed) and sequenced on the same flow cell lane to reduce technical noise. Each PCR reaction used 4 μL of indexing oligonucleotides, 4ul of PCR primer mix, and 2 μL of Multiplex 5X PCR Master Mix. Thermocycling conditions were 37°C for 30 min, 50°C for 10 min, 95°C for 3 min; 18 cycles of 95°C for 15 s, 65°C for 1 min, 68°C for 30 s; and 68°C for 5 min. PCR reactions were pooled and purified twice using AMPure XP beads (Beckman Coulter, A63881), according to the manufacturer’s protocol. Pooled libraries were sequenced at 2 × 27 base pairs with the dual-indexing workflow on an Illumina NovaSeq to generate ~1.1 billion raw reads.
Validation
To validate the spatial transcriptomic results, in situ hybridization images were obtained from the Allen Brain Institute (http://mouse.brain-map.org), converted to gray scale and adjusted for brightness and contrast using Adobe Photoshop (version 23.4.2); all adjustments were applied to the entire image. Criteria for selection of representative in situ hybridization images were 1) previously published mRNAs demonstrating striatal spatiomolecular differential expression including Cnr1, Coch, Col6a1, Cpne6, Crym, Cxcl14, Gpr139, Gpr155, Nefm and Syt1010,12 were excluded to avoid redundancy; 2) FC > 2 between DM and VL for both dSPNs and iSPNs; 3) availability of in situ hybridization images in the coronal plane. Sixteen genes met these criteria including Acvrl1, Astn2, Dpp10, Efna5, Gda, Hpcal4, Islr2, Kcnk2, Kctd8, Me2, Nrgn, Rgs4, Rgs7bp, Sema 7a, Slc41a1, Zbtb20. Four genes were excluded due to the lack of a discernible (over- or under-exposed) and/or specific in situ hybridization signal (Efna5, Islr2, Nrgn, Slc41a1). For the remaining genes, the distribution of the hybridization in the Allen Brain images was consistent with the spatial expression reported here.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data processing
Raw data were processed according to the GeoMx Next Generation Sequencing Pipeline. First, raw reads (FASTQ files) were screened for high Q30 scores, the adapters were removed (resulting in trimmed reads), and the paired-end reads were merged (resulting in stitched reads). Next, the reads were aligned to the readout tag sequence identifier, which identifies each photocleaved oligonucleotide. Then, PCR duplicates were removed by matching on the UMI, resulting in deduplicated reads. These deduplicated reads were then imported into the GeoMx Digital Spatial Profiler Data Analysis Suite for downstream quality control (QC) and statistical analyses.
Quality control
There were two steps in the QC process: Segment QC and Biological Probe QC. The following settings were used for segment QC: number of raw reads ≥1000, percent of aligned reads ≥80%, percent of stitched reads ≥80%, percent of trimmed reads ≥80%, percent sequencing saturation ≥50%, negative probe count geomean ≥10, no template control count <1000, surface area ≥16000, nuclei count ≥200. Next, biological probe QC was performed to determine the threshold for excluding probes that appear to be outliers in the data. Probes were excluded from downstream analyses if the average counts across all segments were ≤10% of the number of probes for that gene (removes probes that perform poorly relative to other probes for the same target gene).
Target genes were excluded if their expression was below a specified threshold in ≥1% of segments. The threshold for exclusion was the limit of quantitation (LOQ), with a minimum value = 2. The LOQ is the negative probes (NegProbes) geometric mean (GeoMean) multiplied by the square of the geometric standard deviation (GeoStdev) of the negative probes: LOQ = GeoMean(|NegProbes -n|) * GeoStdev(NegProbes -n),2 and serves as a threshold for high confidence detection. Finally, data were normalized across all samples. First, expression at the third quartile (Q3) was identified for each sample and the group geometric mean was calculated across all Q3 values. A normalization factor was calculated for each sample by dividing the Q3 value of each sample by the group geometric mean. Expression values for each sample were divided by the normalization factor for that sample. Thus, all samples were normalized so that their Q3s were at the same level, which reduces spurious effects of outliers and broadly accounts for differences in cell density.
Data analysis
For all analyses, the sample size was six mice (n = 6) with three striatal regions (DM, DL, VL) per mouse. Linear Mixed Models with Benjamini-Hochberg correction for multiple comparisons were used to analyze data with repeated measurements from the same animal (Results and Figure 2). Unpaired t tests with Benjamini-Hochberg correction for multiple comparisons were used to analyze data with only one measurement per animal (comparisons of dSPN to iSPN gene expression; Figure 1E and Results). A Benjamini-Hochberg adjusted p value (p-adj) < 0.1 was used to determine differentially expressed genes between SPN subtypes (Figure 1E) to maintain consistency with previously published results.47 A fold change (FC) cut-off of 1.5 with a Benjamini-Hochberg adjusted p value (p-adj) < 0.05 was used to determine differentially expressed genes among striatal regions (Results and Figures 2 and 3). For spatial deconvolution, relative proportions of D1 and A2a cell types associated with each AOI were obtained using the SpatialDecon v1.8.0 R package (Figure S1).48 To deconvolute the D1 and A2a spatial gene expression data, normalized gene expression data was used along with the Adult-Brain_MCA cell profile matrix as cell markers. Background values were computed using the negative probe expression values. The ShinyGO v0.76 application (http://bioinformatics.sdstate.edu/go/) was used for Gene Ontology (GO) enrichment analysis of differentially expressed genes (Figure 4).42 ShinyGO uses a hypergeometric distribution over-representation test to calculate the p value for gene set overlaps. The enriched GO terms were then visualized using GO-Figure!, an open-source software for producing semantic similarity scatterplots.44 A semantic similarity threshold of 0.5 was used for redundancy reduction. Heatmaps were created using R package ComplexHeatmap with default parameters (Figures 4 and S2).43 Data from individual genes were graphed as box and whiskers plots with the box extending from the 25th to the 75th percentiles and a horizontal line at the median (Figure 3; GraphPad Prism version 9.4.1). ANOVAs were performed using GraphPad Prism version 9.4. One-way repeated measures ANOVAs were used to compare cell densities across regions (Results). Two-way repeated measures ANOVAs with region as the repeated measure were used to determine if the number of genes detected varied by region or cell type (Figure 1D legend) and to determine if gene expression was regulated in parallel between dSPNs and iSPNs (Table S2). Tests in Table S2 included only genes that were differentially expressed (FC > 1.5, p-adj <0.05) across striatal subregions in one SPN subtype, but not in the other SPN subtype with FC < 1.25 or in the opposite direction (region x cell type interaction effect). For all ANOVAs, QQ plots (normality distribution plots) were nearly linear and approximated the line of identity.
Supplementary Material
Highlights.
Extensive heterogeneity in spiny projection neuron gene expression among striatal subregions
Direct and indirect spiny projection neurons share molecular signatures across subregions
Regional variation in the expression of genes that mediate neurotransmission
ACKNOWLEDGMENTS
This work was funded by grants from the National Institutes of Health R21 NS114882 (E.J.H.) and R01 NS124764 (E.J.H.) and through the generous support of a Technology Catalyst Award from the Department of Pharmacology and Chemical Biology, Emory University School of Medicine. The Graphical Abstract was created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113435.
DECLARATION OF INTERESTS
A.V. and P.M.R. are former employees of NanoString, and D.K. is currently employed at NanoString. A.V., P.M.R., and D.K. are current shareholders of NanoString.
REFERENCES
- 1.Yin HH, Ostlund SB, Knowlton BJ, and Balleine BW (2005). The role of the dorsomedial striatum in instrumental conditioning. Eur. J. Neurosci 22, 513–523. 10.1111/j.1460-9568.2005.04218.x. [DOI] [PubMed] [Google Scholar]
- 2.Devan BD, and White NM (1999). Parallel information processing in the dorsal striatum: relation to hippocampal function. J. Neurosci 19, 2789–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGeorge AJ, and Faull RL (1989). The organization of the projection from the cerebral cortex to the striatum in the rat. Neuroscience 29, 503–537. [DOI] [PubMed] [Google Scholar]
- 4.Hintiryan H, Foster NN, Bowman I, Bay M, Song MY, Gou L, Yamashita S, Bienkowski MS, Zingg B, Zhu M, et al. (2016). The mouse cortico-striatal projectome. Nat. Neurosci 19, 1100–1114. 10.1038/nn.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, and Mao T (2016). A comprehensive excitatory input map of the striatum reveals novel functional organization. Elife 5, e19103. 10.7554/eLife.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hjorth JJJ, Kozlov A, Carannante I, Frost Nylén J, Lindroos R, Johansson Y, Tokarska A, Dorst MC, Suryanarayana SM, Silberberg G, et al. (2020). The microcircuits of striatum in silico. Proc. Natl. Acad. Sci. USA 117, 9554–9565. 10.1073/pnas.2000671117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerfen CR, and Surmeier DJ (2011). Modulation of striatal projection systems by dopamine. Annu. Rev. Neurosci 34, 441–466. 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong MR, and Wichmann T (2007). Circuits and circuit disorders of the basal ganglia. Arch. Neurol 64, 20–24. 10.1001/arch-neur.64.1.20. [DOI] [PubMed] [Google Scholar]
- 9.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin A, Calvigioni D, Tzortzi O, Fuzik J, Warnberg E, and Meletis K (2019). A Spatiomolecular Map of the Striatum. Cell Rep. 29, 4320–4333.e4325. 10.1016/j.celrep.2019.11.096. [DOI] [PubMed] [Google Scholar]
- 11.Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, and Quake SR (2016). Cellular Taxonomy of the Mouse Striatum as Revealed by Single-Cell RNA-Seq. Cell Rep. 16, 1126–1137. 10.1016/j.celrep.2016.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley G, Gokce O, Malenka RC, Südhof TC, and Quake SR (2020). Continuous and Discrete Neuron Types of the Adult Murine Striatum. Neuron 105, 688–699.e8. 10.1016/j.neuron.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merritt CR, Ong GT, Church SE, Barker K, Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, et al. (2020). Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol 38, 586–599. 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- 15.Wilson CJ, and Groves PM (1981). Spontaneous firing patterns of identified spiny neurons in the rat neostriatum. Brain Res. 220, 67–80. 10.1016/0006-8993(81)90211-0. [DOI] [PubMed] [Google Scholar]
- 16.Alegre-Cortés J, Sáez M, Montanari R, and Reig R (2021). Medium spiny neurons activity reveals the discrete segregation of mouse dorsal striatum. Elife 10, e60580. 10.7554/eLife.60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson CJ, and Kawaguchi Y (1996). The origins of two-state spontaneous membrane potential fluctuations of neostriatal spiny neurons. J. Neurosci 16, 2397–2410. 10.1523/jneurosci.16-07-02397.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plotkin JL, Day M, and Surmeier DJ (2011). Synaptically driven state transitions in distal dendrites of striatal spiny neurons. Nat. Neurosci 14, 881–888. 10.1038/nn.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdeman GL, Ronesi J, and Lovinger DM (2002). Postsynaptic endocannabinoid release is critical to long-term depression in the striatum. Nat. Neurosci 5, 446–451. 10.1038/nn832. [DOI] [PubMed] [Google Scholar]
- 20.Shen W, Plotkin JL, Francardo V, Ko WKD, Xie Z, Li Q, Fieblinger T, Wess J, Neubig RR, Lindsley CW, et al. (2015). M4 Muscarinic Receptor Signaling Ameliorates Striatal Plasticity Deficits in Models of L-DOPA-Induced Dyskinesia. Neuron 88, 762–773. 10.1016/j.neuron.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calabresi P, Maj R, Pisani A, Mercuri NB, and Bernardi G (1992). Long-term synaptic depression in the striatum: physiological and pharmacological characterization. J. Neurosci 12, 4224–4233. 10.1523/jneurosci.12-11-04224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabresi P, Pisani A, Mercuri NB, and Bernardi G (1992). Long-term Potentiation in the Striatum is Unmasked by Removing the Voltage-dependent Magnesium Block of NMDA Receptor Channels. Eur. J. Neurosci 4, 929–935. 10.1111/j.1460-9568.1992.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 23.Braz BY, Belforte JE, Murer MG, and Galiñanes GL (2017). Properties of the corticostriatal long term depression induced by medial prefrontal cortex high frequency stimulation in vivo. Neuropharmacology 121, 278–286. 10.1016/j.neuropharm.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Lovinger DM (2010). Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology 58, 951–961. 10.1016/j.neuropharm.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi S, and Lovinger DM (1997). Decreased probability of neurotransmitter release underlies striatal long-term depression and postnatal development of corticostriatal synapses. Proc. Natl. Acad. Sci. USA 94, 2665–2670. 10.1073/pnas.94.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornito A, Harrison BJ, Goodby E, Dean A, Ooi C, Nathan PJ, Lennox BR, Jones PB, Suckling J, and Bullmore ET (2013). Functional dysconnectivity of corticostriatal circuitry as a risk phenotype for psychosis. JAMA Psychiatr. 70, 1143–1151. 10.1001/jamapsychiatry.2013.1976. [DOI] [PubMed] [Google Scholar]
- 27.Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen CY, Dennison CA, Hall LS, et al. (2022). Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508. 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, Bass N, Bigdeli TB, Breen G, Bromet EJ, et al. (2022). Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516. 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, and Richardson EP Jr. (1985). Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol 44, 559–577. 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Roman KM, Briscione MA, Donsante Y, Ingram J, Fan X, Bernhard D, Campbell SA, Downs AM, Gutman D, Sardar TA, et al. (2023). Striatal Subregion-selective Dysregulated Dopamine Receptor-mediated Intracellular Signaling in a Model of DOPA-responsive Dystonia. Neuroscience 517, 37–49. 10.1016/j.neuroscience.2023.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz B, Fritz BM, Yin F, and Atwood BK (2018). Alcohol exposure disrupts mu opioid receptor-mediated long-term depression at insular cortex inputs to dorsolateral striatum. Nat. Commun 9, 1318. 10.1038/s41467-018-03683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs RA, Branham RK, and See RE (2006). Different neural substrates mediate cocaine seeking after abstinence versus extinction training: a critical role for the dorsolateral caudate-putamen. J. Neurosci 26, 3584–3588. 10.1523/JNEUROSCI.5146-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Li Y, Zhu N, Brimijoin S, and Sui N (2013). Roles of dopaminergic innervation of nucleus accumbens shell and dorsolateral caudate-putamen in cue-induced morphine seeking after prolonged abstinence and the underlying D1- and D2-like receptor mechanisms in rats. J. Psychopharmacol 27, 181–191. 10.1177/0269881112466181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Licheri V, Eckernäs D, Bergquist F, Ericson M, and Adermark L (2020). Nicotine-induced neuroplasticity in striatum is subregion-specific and reversed by motor training on the rotarod. Addict. Biol 25, e12757. 10.1111/adb.12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker LC, Berizzi AE, Chen NA, Rueda P, Perreau VM, Huckstep K, Srisontiyakul J, Govitrapong P, Xiaojian J, Lindsley CW, et al. (2020). Acetylcholine Muscarinic M(4) Receptors as a Therapeutic Target for Alcohol Use Disorder: Converging Evidence From Humans and Rodents. Biol. Psychiatry 88, 898–909. 10.1016/j.bio-psych.2020.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montalban E, Giralt A, Taing L, Schut EHS, Supiot LF, Castell L, Nakamura Y, de Pins B, Pelosi A, Goutebroze L, et al. (2022). Translational profiling of mouse dopaminoceptive neurons reveals region-specific gene expression, exon usage, and striatal prostaglandin E2 modulatory effects. Mol. Psychiatry 27, 2068–2079. 10.1038/s41380-022-01439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puighermanal E, Castell L, Esteve-Codina A, Melser S, Kaganovsky K, Zussy C, Boubaker-Vitre J, Gut M, Rialle S, Kellendonk C, et al. (2020). Functional and molecular heterogeneity of D2R neurons along dorsal ventral axis in the striatum. Nat. Commun 11, 1957. 10.1038/s41467-020-15716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, and Gilad Y (2015). Genomic variation. Impact of regulatory variation from RNA to protein. Science 347, 664–667. 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herkenham M, Lynn AB, Johnson MR, Melvin LS, de Costa BR, and Rice KC (1991). Characterization and localization of cannabinoid receptors in rat brain: a quantitative in vitro autoradiographic study. J. Neurosci 11, 563–583. 10.1523/JNEUROSCI.11-02-00563.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strittmatter SM, Lo MM, Javitch JA, and Snyder SH (1984). Autoradiographic visualization of angiotensin-converting enzyme in rat brain with [3H]captopril: localization to a striatonigral pathway. Proc. Natl. Acad. Sci. USA 81, 1599–1603. 10.1073/pnas.81.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paletzki RF (2002). Cloning and characterization of guanine deaminase from mouse and rat brain. Neuroscience 109, 15–26. 10.1016/s0306-4522(01)00352-9. [DOI] [PubMed] [Google Scholar]
- 42.Ge SX, Jung D, and Yao R (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 44.Reijnders MJMF, and Waterhouse RM (2021). Summary Visualizations of Gene Ontology Terms With GO-Figure. Front. Bioinform 1, 638255. 10.3389/fbinf.2021.638255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Briscione MA, Dinasarapu AR, Bagchi P, Donsante Y, Roman KM, Downs AM, Fan X, Hoehner J, Jinnah HA, and Hess EJ (2021). Differential expression of striatal proteins in a mouse model of DOPA-responsive dystonia reveals shared mechanisms among dystonic disorders. Mol. Genet. Metab 133, 352–361. 10.1016/j.ymgme.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose SJ, Yu XY, Heinzer AK, Harrast P, Fan X, Raike RS, Thompson VB, Pare JF, Weinshenker D, Smith Y, et al. (2015). A new knock-in mouse model of l-DOPA-responsive dystonia. Brain 138, 2987–3002. 10.1093/brain/awv212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748. 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danaher P, Kim Y, Nelson B, Griswold M, Yang Z, Piazza E, and Beechem JM (2022). Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat. Commun 13, 385. 10.1038/s41467-022-28020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unprocessed (raw) Digital Spatial Profiling data are publicly available as of the date of publication. The DOI is listed in the key resources table. The entire normalized dataset is available in the supplemental material.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Tyramide (TSA) linked Opal620 | PerkinElmer | FP1495001KT |
| Cyanine 5 dyes | PerkinElmer | NEL745001KT |
| Neutral Buffer Formalin | EMS Diasum | 15740-04 |
| Syto 83 | ThermoFisher | S11364 |
| Critical commercial assays | ||
| Mm-Adora2a-C2 | ACD | 409438-C2 |
| Mm-Drd1-C3 | ACD | 461908-C3 |
| Mouse Whole Transcriptome Atlas RNA detection probes | Nanostring | GMX-RNA-NGSMsWTA-4 |
| Buffer R | Nanostring | N/A |
| Buffer W | Nanostring | N/A |
| GeoMx Seq Code kit | Nanostring | GMX-NGS-SEQ |
| AmPure XP beads | Beckman Coulter | A63881 |
| Multiplex 5X PCR Master Mix | Nanostring | N/A |
| Deposited data | ||
| Unprocessed (raw) data | This paper | https://doi.org/10.15139/S3/HS5BHE |
| Normalized data | This paper; supplement | N/A |
| In situ hybridization images | Allen Brain Atlas | https://mouse.brain-map.org/static/atlas |
| Experimental models: Organisms/strains | ||
| Mus Musculus C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mus Musculus DBA/2J | The Jackson Laboratory | RRID:IMSR_JAX:000671 |
| Software and algorithms | ||
| ShinyGO v0.76 Web App | (Ge et al.)42 | http://bioinformatics.sdstate.edu/go/ |
| ComplexHeatmap v2.14.0 R package | (Gu et al.)43 | https://doi.org/10.18129/B9.bioc.ComplexHeatmap |
| GO-Figure! Python scripts | (Reijnders and Waterhouse)44 | https://gitlab.com/evogenlab/GO-Figure |
| GraphPad Prism v9.4.1 | GraphPad Software, LLC | https://www.graphpad.com/ |
| GeoMx Next Generation Sequencing Pipeline | Nanostring | N/A |
| GeoMx Digital Spatial Profiler Data Analysis Suite | Nanostring | N/A |
| SpatialDecon v1.8.0 R package | Nanostring | https://github.com/Nanostring-Biostats |
| Other | ||
| Bond RX Fully Automated Research Stainer | Leica Biosystems | N/A |
| Hyb EZ II hybridization oven | ACD | 321710/321720 |
| HybriSlip hybridization covers | Grace Bio-Labs | 714022 |
| Illumina NovaSeq | Novogene | N/A |


