Extended Data 9. Examples of modeled atomic structure by CryoREAD for experimental maps without full atomic structures.
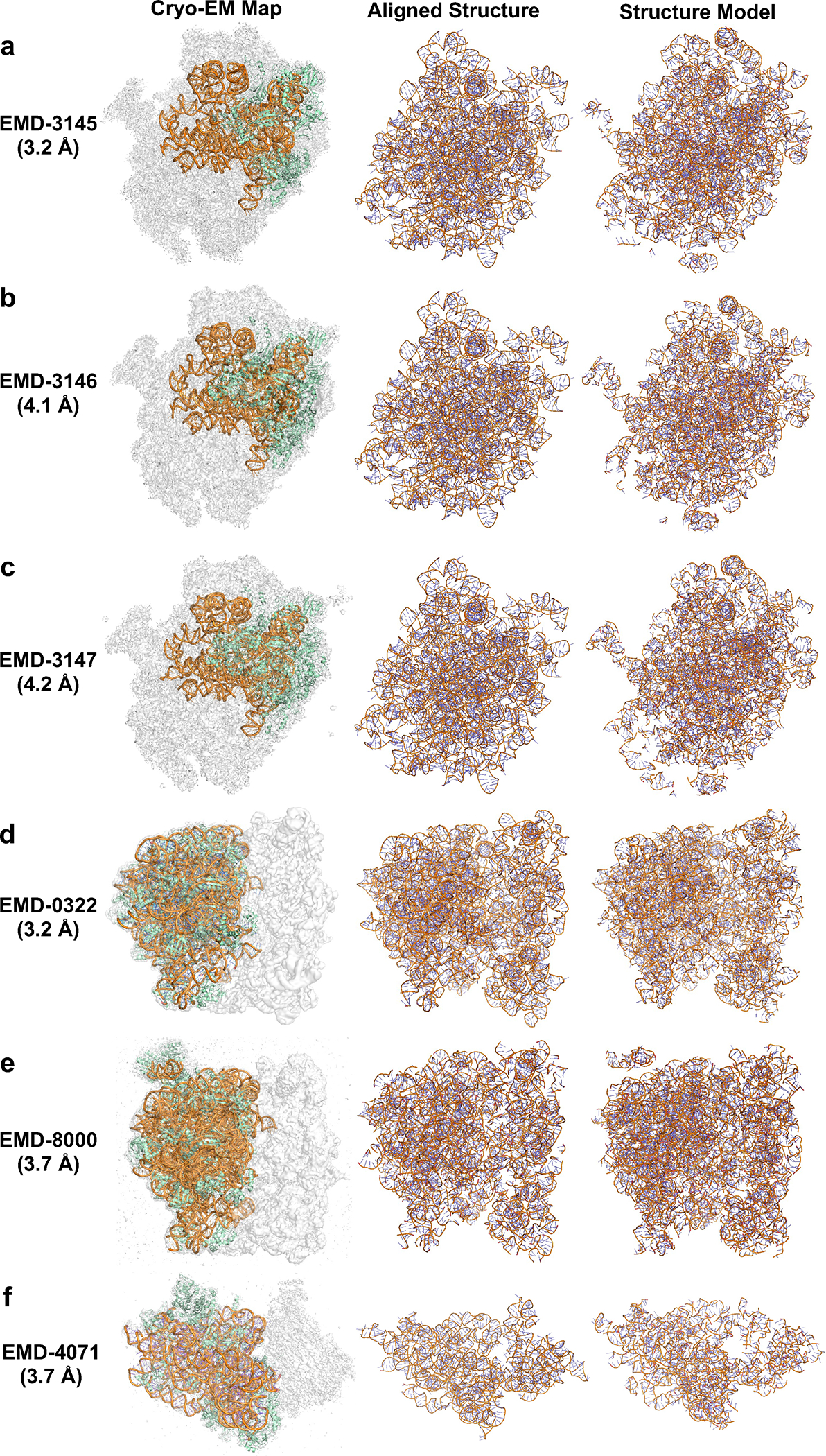
Detailed Evaluation Results are shown in Supplementary Table 3. In this figure, from left to right, the 3 columns correspond to 1) EM map and its corresponding structure; 2) showing only RNA structures in the map. In addition to the RNA structure modeled by the authors, we also shown here homologous structures of missing RNAs in the map, which we searched by BLAST 1. 3) the atomic structure model by CryoREAD. a. the initial Shwachman-Bodian-Diamond syndrome (SBDS) protein closed state of the nascent 60S ribosomal subunit (EMD-3145, PDB-ID: 5AN9, Resolution: 3.3 Å; protein lengths: 1905 aa; RNA length: 1162 nt): Backbone recall: 0.888; Sequence match: 0.696. Identified homologous structure (PDB-ID: 5XXB, RNA length: 3352 nt, Sequence Identity: 84.3%, RMSD: 1.2 Å): Backbone recall: 0.832. b. the SBDS open state of the nascent 60S ribosomal subunit (EMD-3146, PDB-ID: 5ANB, Resolution: 4.1 Å; protein lengths: 3025 aa; RNA length: 1162 nt): Backbone recall: 0.871; Sequence match: 0.544. Identified homologous RNA structure (PDB-ID: 5XXB, RNA length: 3352 nt, Sequence Identity: 84.3%, RMSD: 1.3 Å): Backbone recall: 0.821. c. the ELF1 accommodated state of the nascent 60S ribosomal subunit (EMD-3147, PDB-ID: 5ANC, Resolution: 4.2 Å; protein lengths: 2801 aa; RNA length: 1162 nt): Backbone recall: 0.881; Sequence match: 0.535. Identified homologous structure (PDB-ID: 5XXB, RNA length: 3352 nt, Sequence Identity: 84.3%, RMSD: 1.3 Å): Backbone recall: 0.804. d. TnaC-stalled ribosome complex with the titin I27 domain folding close to the ribosomal exit tunnel (EMD-0322, PDB-ID: 6I0Y, Resolution: 3.2 Å; protein lengths: 3552 aa; RNA length: 3049 nt): Backbone recall: 0.901; Sequence match: 0.632. Identified homologous RNA structure (PDB-ID: 7D80, RNA length: 4761 nt, Sequence Identity: 100%, RMSD: 1.2 Å): Backbone recall: 0.834. e. RNC-SRP-SR complex early state (EMD-8000, PDB-ID: 5GAD, Resolution: 3.7 Å; protein lengths: 4087 aa; RNA length: 3049 nt): Backbone recall: 0.914; Sequence match: 0.655. Identified homologous structure (PDB-ID: 7D80, RNA length: 4761 nt, Sequence Identity: 100%, RMSD: 1.0 Å): Backbone recall: 0.847. f. Structure of the 40S ABCE1 post-splitting complex (EMD-4071, PDB-ID: 5LL6, Resolution: 3.9 Å; protein lengths: 3429 aa; RNA length: 1325 nt): Backbone recall: 0.856; Sequence match: 0.586. Identified homologous structure (PDB-ID: 7OSM, RNA length: 1740 nt, Sequence Identity: 99%, RMSD: 1.0 Å): Backbone recall: 0.784.
In Extended Data 9, we show cases where only a part of the structures in an EM map was modelled by authors. They are maps from three different sets of EM maps of ribosomal subunits. The first set (panel a-c) includes three different states of eIF6 release from the nascent 60S ribosomal subunit 2 of Dictyostelium discoideum, where only part of 26S ribosomal unit is modeled by the authors. The second set (panel d-e) presents two different forms of 70S ribosomal subunit 3 of Escherichia coli, where the authors only modeled 50S ribosomal subunit but 30S ribosomal subunit was left unmodelled. The third example (panel f) is 40S ribosomal subunit of Saccharomyces cerevisiae, where only part of 18S ribosomal RNA was modeled. We filled the missing RNA structure in the maps with homologous RNA structure found by BLAST 1 against PDB. Sequence identities of the identified RNAs were 84.3% to 100%. CryoREAD models for the missing RNA structures had backbone recall of 0.784 to 0.847, when the homologous structures were considered as reference. Backbone recall of CryoREAD models for RNAs with authors’ model was from 0.856 to 0.914.
