Fig. 4. Atomic structure modeling by CryoREAD for experimental maps from SARS-Cov-2 benchmark.
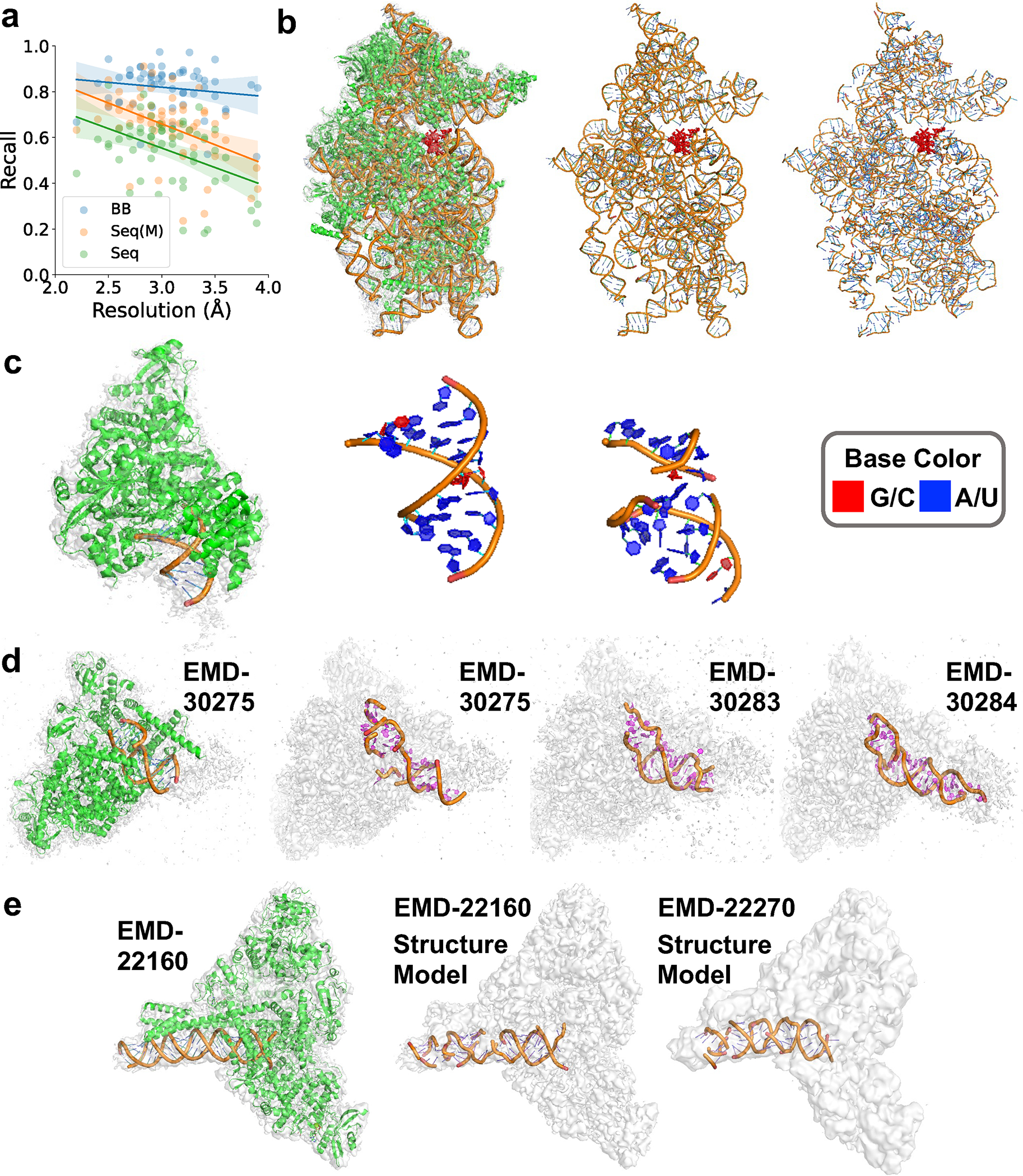
a. the backbone recall (BB), sequence recall (Seq), and sequence match (Seq (M)) relative to the map resolution. Modeling results for the 58 maps that have associated structures were used. For BB, the equation of regression line is (Pearson correlation coefficient: −0.157, p-value: 0.239, standard error:0.035). For Seq, the equation of regression line is (Pearson correlation coefficient: −0.425, p-value: 0.001, standard error:0.048). For Seq(M), the equation of regression line is (Pearson correlation coefficient: −0.460, p-value: 2.844e-4, standard error:0.047). The results of individual maps are provided in Supplementary Table 5_with sequence after refine. b. SARS-CoV-2-Nsp1-40S complex (EMD-11320, PDB ID: 6ZOJ). Resolution: 2.8 Å; protein lengths: 4914 aa; RNA length: 1704 nt: Backbone recall: 0.909; Sequence match: 0.678; Sequence recall: 0.640. From left to right, the EM map and the corresponding structure; only RNA structure in the PDB entry; the atomic structure model by CryoREAD. NSP1 of SARS-CoV-2 is shown in red. c. Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP (EMD-11692, PDB ID: 7AAP). Resolution: 2.5 Å; protein lengths: 1120 aa; RNA length: 21 nt. Backbone recall: 0.773; Sequence match: 0.75; Sequence recall: 0.571. d. COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex (EMD-30275, PDB-ID:7C2K). Resolution: 2.93 Å; protein lengths: 1256 aa; RNA length: 31 nt. Backbone recall: 0.831; Sequence match: 0.630; Sequence recall: 0.548. We built models for maps in two independent conformations, the conformation I (EMD-30283, Resolution: 3.03 Å) and the conformation II (EMD-30284, Resolution: 3.12 Å). We used COOT to refine the structure models by CryoREAD. e. SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (EMD-22160, PDB ID: 6XEZ). Resolution: 3.5 Å; protein lengths: 2563 aa; RNA length: 70 nt. Backbone recall: 0.844; Sequence match: 0.613; Sequence recall: 0.543. On the right is the RNA model by CryoREAD for SARS-CoV-2 replication/transcription complex bound to nsp13 helicase - nsp13(1)-RTC (EMD-22270, Resolution: 4.0 Å). No structure model was associated with this map.
