Abstract
Objectives
To identify indolepropionate (IPA)-predicting gut microbiota species, investigate potential diet-microbiota interactions, and examine the prospective associations of circulating IPA concentrations with type 2 diabetes (T2D) and coronary heart disease (CHD) risk in free-living individuals.
Design
We included 287 men from the Men’s Lifestyle Validation Study, a sub-study of the Health Professionals Follow-Up Study (HPFS), who provided up to two pairs of fecal samples and two blood samples. Diet was assessed using 7-day diet records. Associations between plasma concentrations of tryptophan metabolites and T2D CHD risk were examined in 13,032 participants from Nurses’ Health Study (NHS), NHSII, and HPFS.
Results
We identified 17 microbial species whose abundance was significantly associated with plasma IPA concentrations. A significant association between higher tryptophan intake and higher IPA concentrations was only observed among men who had higher fiber intake and a higher microbial species score consisting of the 17 species (p-interaction < 0.01). Dietary and plasma concentrations of tryptophan and most kynurenine pathway metabolites were positively associated with T2D risk (HRQ5 vs Q1 ranged from 1.17 to 1.46) while a significant inverse association was found for IPA (HRQ5 vs Q1 [95% CI] = 0.70 [0.56, 0.88]). No associations were found in CHD for any plasma tryptophan metabolites.
Conclusions
Specific microbial species and dietary fiber jointly predicted significantly higher circulating IPA concentrations at higher tryptophan intake. Dietary and plasma tryptophan, as well as its kynurenine pathway metabolites demonstrated divergent associations from those for IPA, which was significantly predictive of lower risk of T2D.
INTRODUCTION
Tryptophan is an essential amino acid that plays critical roles in human nutrition and metabolism. Ingested tryptophan is predominantly catabolized through the kynurenine pathway (KP) in the liver and peripheral tissues, while a small fraction that is not absorbed can be directly utilized by the gut microbiota to produce indole and its derivatives.(1) Interestingly, the host and microbial tryptophan catabolites appear to manifest divergent associations with human health. While the increased level of KP catabolites such as kynurenine has been implicated in the development of cardiometabolic diseases,(2,3) higher circulating concentrations of microbial tryptophan metabolites, most notably the indolepropionate (IPA), is associated with a lower risk of T2D.(4,5) Since dietary tryptophan is the only source of tryptophan that can be converted to IPA, the gut microbiota profiles are likely to play a pivotal role in modulating the IPA production. Although culture-based studies in animal models have identified a number of IPA-producing microbial species,(6) the associations between IPA and gut microbiota profile in free living-individuals remain to be fully characterized. Recent investigations among human populations have discovered some gut microbial genus and species that may potentially produce IPA but these are quite different from findings from rodent models, implying limited generalizability of host-microbial crosstalk in tryptophan metabolism from animal studies to free-living human individuals.(7,8) In addition, emerging evidence from population-based studies suggested that dietary fiber, as a key prebiotic, may be involved in the production of IPA since a higher dietary fiber intake has been consistently associated with increased circulation IPA concentrations as well as an overlapping panels of gut microbial species associated with both fiber and IPA.(5,7,and 8) As such, the role of dietary fiber is particularly intriguing as no evidence suggests IPA can be produced through fiber fermentation and the production of IPA was found to be completely gut microbiota-dependent in rodent models.(9) Because no previous population-based study explicitly takes dietary fiber intake into consideration when examining the interactions between dietary tryptophan and gut microbiota profile on circulating IPA concentrations, the potential complex interplay between dietary and gut microbial profiles that modulates the IPA production remains to be elucidated.
The current study aims to integrate the gut microbiome and plasma metabolome data to identify gut microbial species associated with circulating IPA concentrations and explicitly investigate the interaction between diet and gut microbiome with circulating IPA level. We also investigated whether dietary fiber may strengthen the IPA-producing capacity of the identified gut microbiota species. Finally, to complement the host-microbial interaction analysis, prospective associations of dietary tryptophan as well as major tryptophan metabolites with T2D and coronary heart disease (CHD) risk were examined in three large prospective cohort studies of US men and women.
METHODS
Study population
The current study consisted of two primary components of analyses (Figure 1): 1) gut microbial taxonomic and functional analyses that elucidated the relationships among dietary tryptophan and intake, gut microbiome, and plasma concentrations of IPA; 2) survival analyses that examined the prospective associations between circulating level of IPA and other tryptophan metabolites and incidence of T2D. We also considered CHD in this analysis given the etiology of T2D and CHD may partially overlap. The study population for the gut microbiome analyses is the Men’s Lifestyle Validation Study (MLVS), a 1-year sub-study nested in the Health Professionals Follow-up Study (HPFS) which is an ongoing prospective cohort study initiated in 1986 enrolling 51,529 US male health professionals. During 2011-2013, a total of 307 participants from MLVS provided up to two pairs of self-collected fecal samples with each pair collected from two consecutive bowel movements 24 to 72 hours apart; the two pairs were collected approximately 6 months apart. Two blood samples were also collected, 6-month apart, to roughly match with the timing of the stool sample collection. (10) After excluding 20 participants without blood samples, we finally included 287 men in the gut microbiome analyses.
Figure 1.
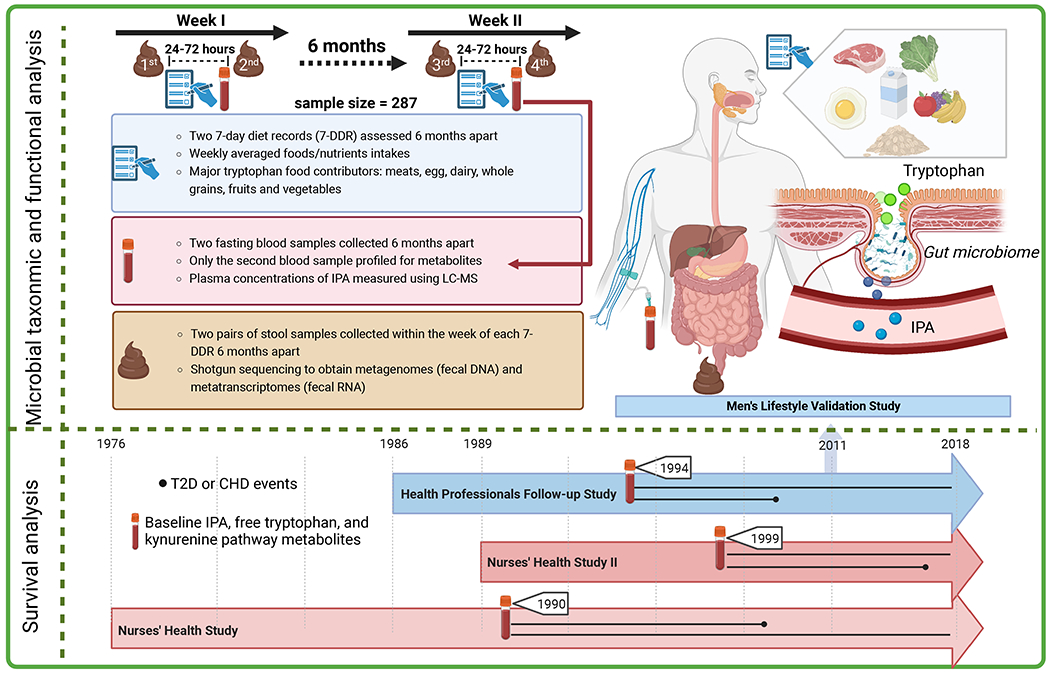
Overview of the dual components of the study design integrating diet, gut microbiota, and circulating metabolites in relation to IPA and T2D. The microbial analysis focused on exploring the inter-relationships among dietary tryptophan, fiber, gut microbial features, and plasma level of IPA. The survival analysis examined the prospective associations of dietary tryptophan and circulating tryptophan metabolites with T2D risk.
The study population of the survival analyses were sub-studies among participants from the Nurses’ Health Study (NHS), NHSII, and HPFS who provided blood samples during 1990-1999 (1990 for NHS, 1999 for NHSII, and 1994 for HPFS). The survival analysis sample consisted of 13,669 participants from the three cohort studies who had data of plasma tryptophan metabolites. Similar to HPFS, the NHS (n=121,700) and NHSII (116,340) are two ongoing prospective cohort studies of middle-aged-to-older female nurses who have been followed up since 1976 and 1989, respectively. In all three cohorts, participants were sent questionnaires biennially to update their demographic and lifestyle information and to identify disease incidence. The cumulative response rates in three cohorts exceeded 90%.(11,12) After excluding prevalent T2D and CHD cases from the 13,669 participants, we included 13,032 participants in the survival analyses.
The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
Taxonomic and functional profiling of stool samples
Stool samples were profiled using Illumina HiSeq paired-end shotgun sequencing to obtain metagenomes and metatranscriptomes. QC was performed according to HMP protocols with KneadData. Detailed information regarding sample processing, sequencing, and QC have been described extensively in previous studies.(13,14) Taxonomic and functional profiles were generated following the bioBakery workflow:(15) MetaPhlAn2 was used to perform taxonomic profiling(16) and HUMAnN2 for functional profiling.(17) For taxonomic features, we excluded those with a relative abundance <10−4 (0.01%) in over 10% of all observations; for functional features, we excluded those with a relative abundance <10−5 (0.001%) in over 10% of all observations. Because no metatranscriptomic data (RNA data from gene expression of gut microbiome) were left after filtering in a subset of 91 men, the microbial analysis only focused on the metagenomic features. We included 925 metagenomes from 287 men in the final analyses.
Metabolomics profiling of blood samples
For both microbiome analyses and survival analyses, the plasma concentrations of tryptophan metabolites including IPA, KP metabolites, and serotonin were profiled in the laboratory at the Broad Institute of Harvard University and M.I.T. (Cambridge, MA), using high-throughput liquid chromatography-tandem mass spectrometry techniques.(18) In MLVS, the mean coefficient of variance (CV) for IPA was 15.4%. In the survival analysis sample, for major tryptophan metabolites, the average CV ranged from 13.7% for indoxyl sulfate to 28.2% for IPA (Supplemental table 1).
Dietary assessment
In the microbiome analyses, diet was assessed using two sets of 7-day diet records (7DDR) administered 6 months apart during the stool sample collection period to record detailed consumptions for seven consecutive days.(19) Provided with a food scale, ruler, and instructional supports via DVD and telephone, participants measured and reported weights in gram of foods before and after eating a meal so that the actual intake could be computed. Moreover, the participants also provided recipes of all home-prepared foods, including the number of servings in each recipe and the portion of the foods they consumed. Over 150 nutrients and dietary constituents including tryptophan and fiber (total fiber, soluble fiber, insoluble fiber, and pectin) were derived from 7DDRs based on the Nutrition Data System for Research at the University of Minnesota Nutrition Coordinating Center.(20) Dietary tryptophan was calculated by multiplying the frequency of consumption of each tryptophan-containing food item by tryptophan content and then summing across from all foods. Since the IPA was only measured in the second blood samples collected in the MLVS, we used average values of food and nutrients intake from two 7DDRs to represent the recent dietary intake contemporaneous with stool sample collection.
In the survival analyses, diet was assessed by a validated FFQ every 4 years starting in 1984 in the NHS, 1991 in the NHSII, and 1986 in the HPFS, respectively. For each food item listed in the FFQ, participants were asked their average consumption frequency of a pre-specified portion size during the previous year.(21) The average daily intake of tryptophan was calculated by multiplying the frequency of consumption of each tryptophan-containing food item by tryptophan content and then summing across from all foods.
Ascertainment of type 2 diabetes and coronary heart disease
Participants who reported a diabetes diagnosis were mailed a supplementary questionnaire regarding symptoms, diagnostic tests, and hypoglycemic therapy. We used the National Diabetes Data Group criteria to ascertain diabetes diagnoses for cases identified before 1998 or the American Diabetes Association criteria for cases after 1998.(22,23) Our validation studies demonstrated about 97% of participants with self-reported T2D confirmed by the questionnaire were reconfirmed by review of the medical records.(24,25)
Total CHD included myocardial infarction (MI) and fatal CHD and we included both definite and probable cases in the analysis. Non-fatal MI was reported by the participants in the follow-up questionnaires and was subsequently ascertained by medical records review. Study physicians who were blinded to exposure status reviewed the medical records and confirmed/refuted reported MI cases according to the WHO criteria.(26,27) Fatal CHD was confirmed by a review of hospital records or autopsy reports if CHD was listed as the underlying cause of death and if evidence of previous CHD was available from medical records.
Assessment of covariates
Biennial follow-up questionnaires were sent to participants in 3 cohorts to collect and update the occurrence of diseases, demographic information, and lifestyle factors including smoking status, alcohol consumption, and body weight, and other variables. Physical activities were repeatedly assessed in the three cohorts and we calculated the body mass index (BMI) as weight in kilograms divided by the square of height in meters to measure overall obesity. In addition, we used the alternative healthy eating index (AHEI) to adjust for overall diet quality.(28) In MLVS, the covariates information was collected in similar ways around the time of blood collection, except that we additionally assessed the information on use of probiotics and antibiotics, surgical procedures, and the Bristol stool chart through fecal sample collection questionnaires.
Statistical analysis
In the microbiome analyses, we calculated the intake of energy-adjusted tryptophan and fiber as well as major tryptophan food contributors using the averages of two 7DDRs to represent a more stable, habitual diet, also partially because IPA and other metabolites were measured in the 2nd blood samples only in the MLVS. Similarly, we averaged up to 4 measurements of metagenomic features from two pairs of fecal samples including microbial taxon, DNA enzymes, and DNA pathways. When linking metagenomes with diet, for participants who did not provide fecal samples in all 4 phases, we used the dietary intake data from the 7DDR that was closest to the fecal sample collections. For example, for participants who only provided fecal samples in phase 1 and 2, we only used their first 7DDR information.
Multiple linear regression was used to assess the associations between dietary tryptophan as well as major tryptophan contributors and plasma IPA concentrations. Bray-Curtis dissimilarity-based principal coordinates (PCo) analysis was conducted using taxonomic data at the species level and the PERNOVA test was performed to assess associations between the overall microbial community and plasma IPA concentrations. After applying Arc-sin square root transformation to relative abundances of taxonomic features, the associations between metagenomic features and plasma concentrations of IPA were analyzed using generalized fixed-effects linear regressions implemented in MaAsLin2. Multivariable models were adjusted for age, BMI, calorie intake (quintile), physical activity (quintile), smoking status (never, past, current), cumulatively averaged AHEI scores, probiotic use, antibiotic use, colonoscopy, acid reduction medication use, and Bristol stool chart (Supplemental figure 1).(10,29, and 30) In addition to IPA, we also analyzed the associations with dietary tryptophan and major tryptophan food contributors including vegetables, legume, nuts, fruits, red/processed meat, poultry, fish, egg, dairy, and whole grain. The functional analyses including microbial enzymes and pathways were analyzed in a similar manner.
To evaluate the modulation effects of gut microbiome on the dietary tryptophan-IPA associations, we derived a species rank score (SRS) based on identified IPA-associated species to reflect the IPA-producing capacity. We ranked the positive IPA-predicting species by their relative abundance of non-zero values. The negative IPA-predicting species were reversely ranked in the same manner. The SRS was the sum of rank from both IPA-predicting and non-predicting species. The interactions between dietary tryptophan intake and gut microbiome on plasma concentration of IPA were examined by including a product term between dietary tryptophan and SRS in a generalized linear regression model. To further consider the role of dietary fibers on modulating the diet-gut microbiome interaction, we additionally introduced in a product term among dietary tryptophan, SRS, and total fiber as well as the pair-wised product terms among these three variables in the multivariable-adjusted model. In a secondary analysis, we also examined the 3-way interactions with subtypes of fiber, including soluble fiber, insoluble fiber, and pectin.
In the survival analyses, plasma tryptophan metabolites with less than 5% of missing values were imputed with the one half of the minimum value and transformed to natural logarithmic Z-score. We cumulatively averaged the dietary tryptophan intake until the blood draw year to represent the baseline tryptophan intake. All tryptophan-related variables were categorized into quintiles. Multivariable Cox proportional hazards model was used to calculate hazard ratios and 95% confidence intervals for the associations between dietary tryptophan and plasma tryptophan metabolites, and the risk of T2D. The proportional hazards assumption was evaluated by including an interaction term between tryptophan intake or plasma tryptophan metabolites and the duration of follow-up and we did not detect any evidence of violations of the assumption (P>0.05 for all tests). The Cox models adjusted for age, race, cohort origin, physical activity (quintile), alcohol intake (quintile), AHEI without alcohol (quintile), smoking status (never, past, current), BMI, and total energy intake (quintile). The median value for each exposure was used to calculate the P value for trend. To plot the multivariable-adjusted Kaplan-Meir survival curves, we used inverse probability weighting method to adjust for covariates.(31) We also conducted a mediation analysis to quantify the mediation effects of KP pathway, IPA, and free plasma tryptophan in the dietary tryptophan-T2D association.(32) Four KP metabolites including kynurenine, kynurenic acid, quinolinic acid, and xanthurenic acid were summed up to represent the KP pathway in the mediation analysis.
In taxonomic and functional analysis, a false discovery rate (FDR)<0.05 were deemed statistically significant. Otherwise, statistical tests were 2-sided with significant level of 0.05. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC) and R v4.1.0.
Patient and public involvement
No patients were involved in setting the research question or the outcome measures, nor were they involved in the design and implementation of the study. No patients were asked to advise on interpretation or writing up of results. We did not have the infrastructure, resources, funding, or time to involve the public in study design, result interpretation, or publication.
RESULTS
Among 287 MLVS participants, higher plasma concentrations of IPA were associated with lower BMI, higher levels of physical activity, overall diet quality, fiber intake, total energy, and lower prevalence of acid reduction medication use (Table 1.1). Other characteristics including age, probiotic use, antibiotic use, colonoscopy, stool types, and dietary tryptophan intake did not appear to be associated with plasma IPA concentrations. Plasma IPA concentrations were modestly correlated with higher consumptions of fiber, vegetables, fruits, legume, nuts, and whole grains while negative correlations were observed for red/processed meat, egg, and dairy (Supplemental figure 2). For the survival analyses, higher baseline plasma IPA levels appeared to be correlated with lower prevalence of overweight/obesity, higher physical activity and overall diet quality level, as well higher plasma concentrations of tryptophan and kynurenine but not other tryptophan metabolites (Table 1.2).
Table 1.1.
Baseline (2011) characteristics of participants in the MLVS according to plasma IPA concentrations.
| Plasma IPA concentration, log Z-score | |||
|---|---|---|---|
|
| |||
| Characteristics | T1 (N=95) | T2 (N=97) | T3 (N=95) |
|
|
|||
| Plasma IPA (log-Z score) | −0.9 (0.7) | 0.1 (0.2) | 0.9 (0.4) |
| Age at fecal sample collection | 71.2 (4.1) | 71.1 (4.1) | 72.2 (4.5) |
| BMI, kg/m2 | 27.6 (4.0) | 25.2 (2.8) | 25.4 (4.2) |
| Physical activity (MET-h/wk) | 114 (58.9) | 115 (51.2) | 122 (42.9) |
| Current smokers, % | 3.2 | 1.0 | 0 |
| Probiotic use, % | 10.5 | 7.2 | 7.4 |
| Antibiotic use, % | 33.7 | 32.0 | 31.6 |
| Acid reduction medication use, % | 30.5 | 19.6 | 17.9 |
| Colonoscopy, % | 8.4 | 4.1 | 4.2 |
| Bristol stool chart, % | |||
| 1-2, hard stool | 6.3 | 2.1 | 5.3 |
| 3-5, normal stool | 58.9 | 66.0 | 66.3 |
| 6-7, loose stool | 11.6 | 7.2 | 8.4 |
| Long-term AHEI score * | 55.5 (8.4) | 58.5 (8.8) | 61.0 (8.7) |
| Dietary tryptophan intake (g/d) | 1.1 (0.2) | 1.0 (0.3) | 1.0 (3) |
| Total fiber intake (g/d) | 23.8 (7.4) | 26.8 (7.5) | 31.8 (12.1) |
| Soluble fiber intake (g/d) | 7.7 (2.6) | 8.3 (2.7) | 9.8 (3.7) |
| Insoluble fiber intake (g/d) | 16.0 (5.5) | 18.4 (5.6) | 21.8 (8.9) |
| Pectin intake (g/d) | 3.2 (1.2) | 3.6 (1.3) | 4.5 (2.3) |
| Total energy (Kcal/d) | 2250 (476) | 2290 (480) | 2320 (463) |
Values are means (SD) for continuous variables or otherwise indicated, or percentages (%) for categorical variables.
Long-term AHEI score was calculated as the cumulative average values from 1986 to 2010.
Table 1.2.
Baseline characteristics of survival analysis participants in the NHS (1990), NHSII (1999), and HPFS (1994) according to plasma IPA concentrations.
| Baseline plasma IPA concentration, log Z-score | |||||
|---|---|---|---|---|---|
|
| |||||
| Characteristics | Q1 (N=1,012) | Q2 (N=1,012) | Q3 (N=1,012) | Q4 (N=1,013) | Q5 (N=1,012) |
|
|
|||||
| Baseline plasma tryptophan metabolites, log Z-score * | |||||
| IPA | −1.1 (−1.7, −0.9) | −0.4 (−0.5, −0.2) | 0.08 (−0.01,0.2) | 0.5 (0.4,0.6) | 1.1 (0.9,1.4) |
| Tryptophan | −0.08 (−0.8,0.5) | −0.03 (−0.6,0.6) | 0.1 (−0.5,0.6) | 0.08 (−0.4,0.6) | 0.2 (−0.4,0.7) |
| Indoxyl sulfate | −0.07 (−0.7,0.5) | 0.2 (−0.4,0.7) | 0.2 (−0.4,0.7) | 0.2 (−0.4,0.7) | −0.02 (−0.7,0.5) |
| Serotonin | 0.02 (−0.6,0.7) | 0.2 (−0.5,0.8) | 0.10 (−0.6,0.7) | 0.1 (−0.6,0.7) | 0.3 (−0.4,0.8) |
| Kynurenine | −0.04 (−0.7,0.6) | −0.01 (−0.6,0.6) | 0.05 (−0.5,0.6) | 0.1 (−0.4,0.6) | 0.04 (−0.5,0.6) |
| Kynurenic acid | −0.08 (−0.7,0.6) | −0.02 (−0.7,0.7) | 0.06 (−0.5,0.7) | 0.03 (−0.6,0.7) | −0.03 (−0.6,0.6) |
| Quinolinic acid | 0.03 (−0.6,0.7) | −0.06 (−0.6,0.6) | −0.02 (−0.6,0.6) | 0.04 (−0.6,0.6) | −0.08 (−0.7,0.5) |
| Xanthurenic acid | 0.1 (−0.6,0.6) | 0.09 (−0.5,0.6) | 0.1 (−0.4,0.6) | 0.1 (−0.4,0.6) | 0.09 (−0.5,0.6) |
| Dietary tryptophan intake (g/d) | 1 (0.3) | 1 (0.3) | 1 (0.3) | 1 (0.3) | 1 (0.3) |
| Age (years) | 53 (8.9) | 53 (9.6) | 52 (9.7) | 52 (9.3) | 53 (9.8) |
| Ethnicity, % | |||||
| White | 96.7 | 97.5 | 98.7 | 97.2 | 97.2 |
| Black | 1.1 | 1.0 | 0.4 | 0.6 | 0.7 |
| Asian | 1.2 | 0.8 | 0 | 1.2 | 0.7 |
| Other | 1.0 | 0.7 | 0.9 | 1.0 | 1.4 |
| Cohort origins, % | |||||
| NHS | 50.0 | 43.0 | 39.3 | 42.3 | 45.5 |
| NHSII | 37.6 | 43.3 | 47.9 | 47.4 | 40.9 |
| HPFS | 12.4 | 13.7 | 12.7 | 10.3 | 13.6 |
| Smoking status, % | |||||
| Never smokers | 51.2 | 55.7 | 56.0 | 58.6 | 62.4 |
| Past smokers | 17.9 | 10.4 | 10.2 | 6.0 | 5.4 |
| Current smokers | 30.9 | 33.9 | 33.8 | 35.3 | 32.2 |
| BMI, % | |||||
| < 25 kg/m2 | 41.8 | 50.2 | 51.0 | 55.4 | 61.2 |
| 25-29.9 kg/m2 | 34.5 | 34.2 | 33.8 | 29.6 | 27.2 |
| BMI≥30 kg/m2 | 22.3 | 14.1 | 14.3 | 14.2 | 10.4 |
| Physical activity (MET-h/wk) | 8.6 (3.2, 20.3) | 11.2 (4.2, 24.3) | 12.4 (4.4, 25.7) | 12.7 (5.2, 25.9) | 13.9 (4.9, 27.7) |
| Alcohol consumption (g/d) | 1.8 (0, 6.9) | 1.5 (0, 6.9) | 1.1 (0, 6.0) | 1.8 (0, 6.5) | 1.8 (0, 7.9) |
| AHEI | 44.5 (9.9) | 45.6 (10.3) | 46.5 (10.2) | 47.5 (10.2) | 50.1 (11.0) |
| Total energy intake (Kcal/d) | 1800 (466) | 1820 (494) | 1800 (471) | 1830 (481) | 1820 (480) |
Values are means (SD) or median (IQR) for continuous variables and percentages (%) for categorical variables.
The associations of dietary tryptophan and tryptophan food contributors and overall gut microbial community in relation to plasma IPA level are shown in Figure 2. Although higher dietary tryptophan appeared to be associated with lower plasma IPA level (slope=−0.48, p=0.06), some fiber-rich food contributors including fruits (slope= 0.11, p<0.001), vegetables (slope= 0.08, p=0.01), and whole grains (slope=0.06, p=0.02) were significantly associated with higher plasma IPA concentrations while animal product intake manifested mostly inverse associations (Figure 2 Panel A). Based on 139 species from seven phyla spanning three kingdoms, plasma IPA concentration was weakly but statistically significantly associated with overall gut microbial community abundance (Figure 2 Panel B). In the stratified analysis by two PCos, we did not observe any statistically significant difference between higher and lower PCo in relation to plasma IPA level for dietary tryptophan and tryptophan food contributors (Figure 2 Panel C).
Figure 2.
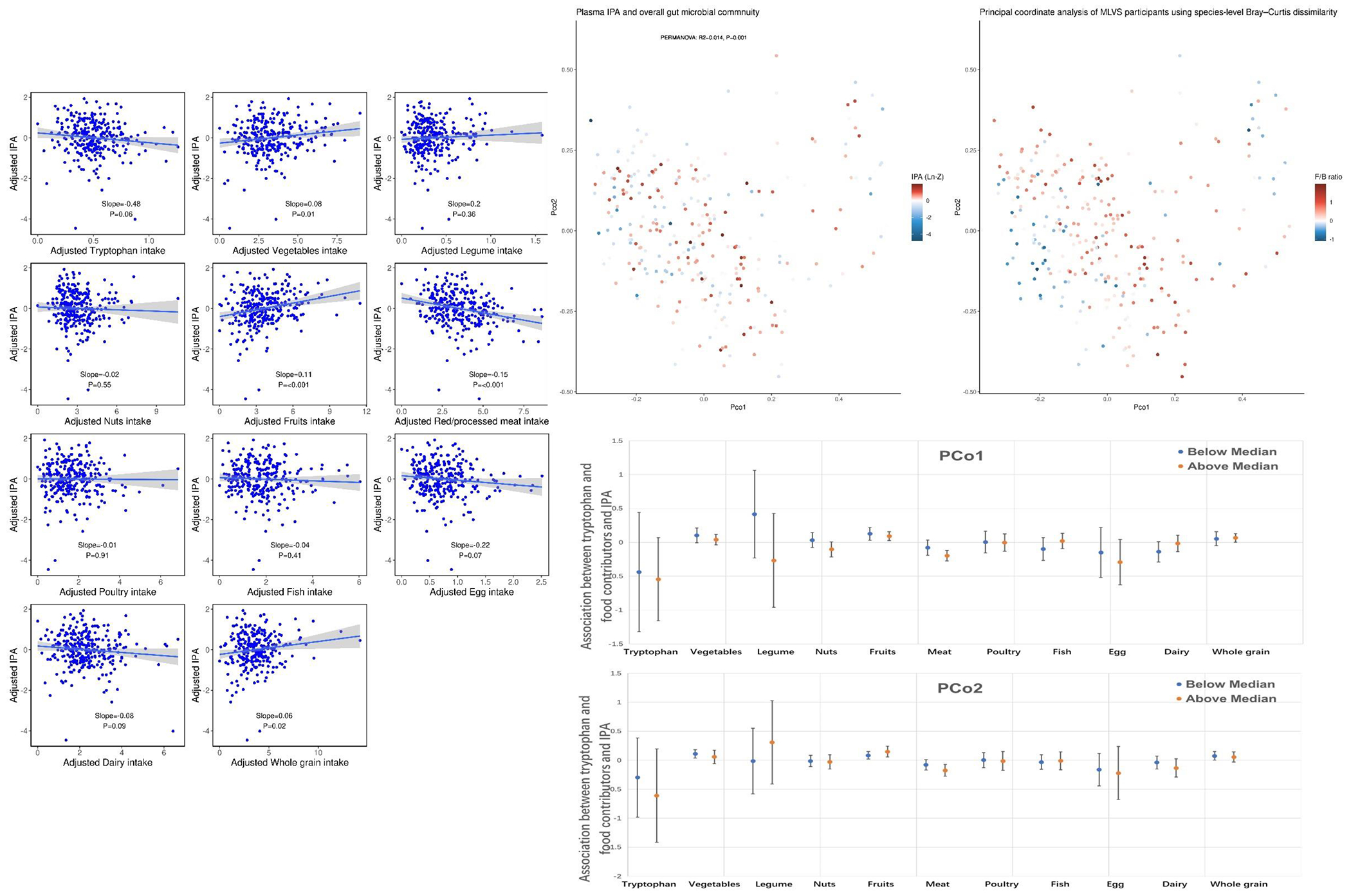
Associations between dietary tryptophan, major tryptophan food contributors, and overall gut microbial community and plasma IPA concentrations.
Panel A: Associations between dietary tryptophan and major tryptophan food contributors on plasma IPA concentrations.
Panel B: The first two principal coordinates (PCo) based on based on Bray-Curtis dissimilarity plotted by plasma IPA levels (left) and Firmicutes to Bacteroidetes ratio (right).
Panel C: Associations between dietary tryptophan and major tryptophan food contributors on plasma IPA concentrations stratified by PCo1 and 2. Error bar represents 95% confidence interval.
In the taxonomic analysis, we identified 17 microbial species in two phyla (Bacteroidetes and Firmicutes) whose relative abundance was significantly associated with plasma IPA level (Figure 3, Panel A). Of 17 identified species, only 3 species were positively associated with IPA concentrations (E. eligens, B. crossotus, and F. prausnitzii) and the rest of species were negatively associated with plasma IPA level. Moreover, dietary tryptophan, most major tryptophan food contributors as well as total dietary fiber shared similar associations with the 17 identified species as with IPA (Figure 3, Panel B).
Figure 3.
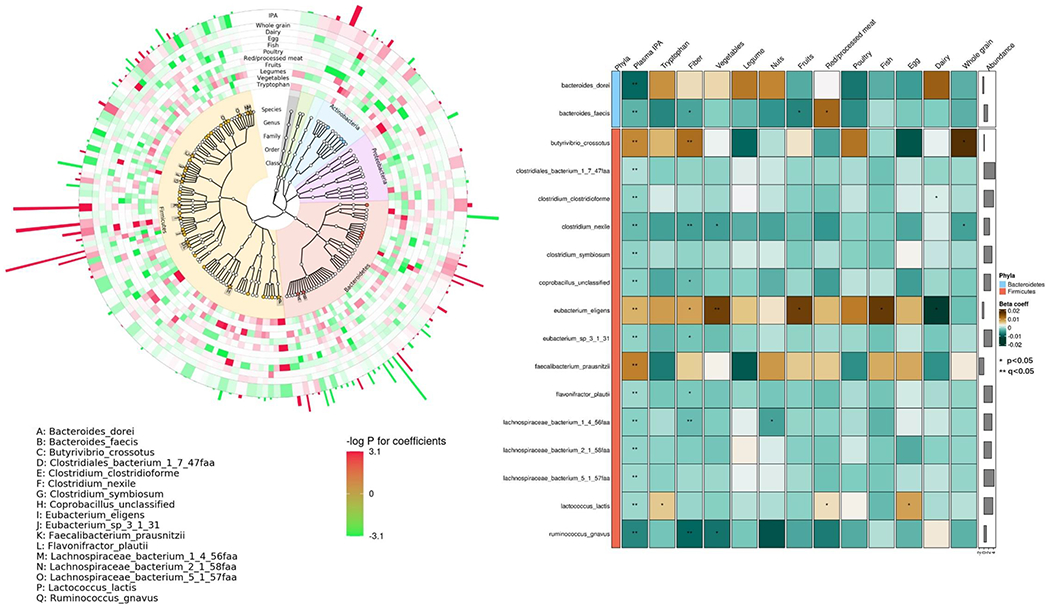
Associations between taxonomic features and plasma IPA concentrations Panel A shows phylogenetic tree for taxonomic features qualified for analysis, highlighting features significantly associated with plasma IPA concentrations (solid stars with letter labels, false discovery rate (FDR)<0.05; solid circles, p<0.05). The inner rings denote associations of each species with IPA levels and intakes of tryptophan food contributors (red, positive associations; green, inverse associations; color depth, statistical significance), and the abundance of each species are portrayed in the outermost ring. Panel B presents the associations between IPA-associated species with dietary tryptophan, fiber, and major tryptophan food contributors. Colors of cells indicate association coefficients, and asterisks denote association significance (** FDR<0.05, *p<0.05). Generalized linear fixed-effects regressions implemented in MaAsLin2 were adjusted for age, BMI, calorie intake (quintile), physical activity (quintile), smoking status (never, past, current), cumulatively averaged AHEI scores, probiotic use, antibiotic use, colonoscopy, acid reduction medication use, and Bristol stool chart.
Higher IPA-producing capacity reflected by a higher SRS did not modify the relationship between dietary tryptophan intake and plasma IPA concentrations (p for interaction = 0.37). Increased tryptophan intake was consistently associated lower plasma IPA level regardless of the SRS (Figure 4, Panel A). We also did not observe any statistically significant interactions with individual IPA-associated species (Supplemental table 2). However, when additionally considering dietary fiber in these interactions, we found a robust, statistically significant interaction between dietary tryptophan and SRS on plasma IPA concentrations at higher level of dietary fiber intake (Figure 4, Panel B, p for 3-way interaction <0.001). Similar significant 3-way interactions were also observed in fiber-rich tryptophan food contributors including vegetables (p=0.002), nuts (p=0.04), and whole grains (p=0.002) while the interactions in the opposite direction were found in red/processed meat (p=0.01), egg (p=0.02), and dairy (p=0.01). Moreover, consistent patterns of interactions were found for all fiber subtypes with different strength of associations (Supplemental table 3).
Figure 4.
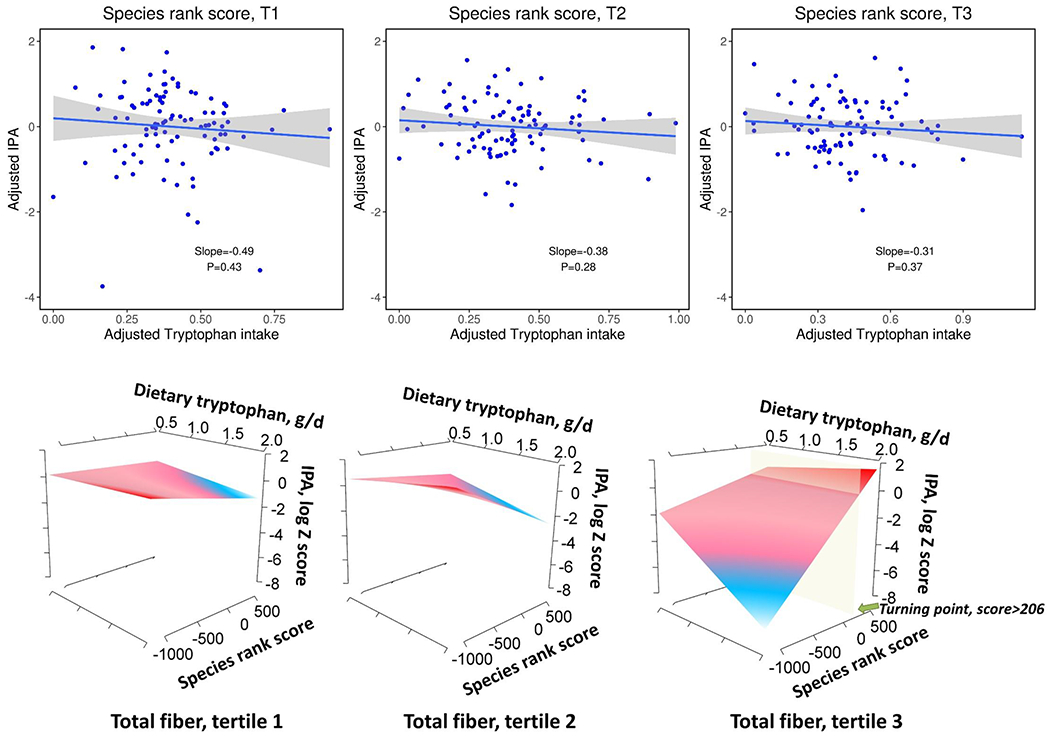
Interactions among dietary tryptophan, species rank score, dietary fiber with plasma concentrations of IPA. Panel A: The associations between dietary tryptophan intake and plasma IPA concentrations according to tertiles of species rank score. Panel B: Interaction between dietary tryptophan and species rank score on plasma IPA concentrations according to tertiles of total fiber intake.
In the functional analysis focusing on the 17 IPA-predicting species, we did not identify any specific enzymes operating in the indole pathway and most statistically significant signals were some unnamed enzymes. Several nucleotides homeostasis-related enzymes were identified in F. prausnitzii (Supplemental figure 2, Panel A) that were significantly associated with the IPA levels. Of note, total fiber and fiber subtypes shared similar associations with identified enzyme signals with plasma IPA. Finally, we identified 54 pathways within 5 species including B. dorei, B. faecis, B. crossotus, F. prausnitzii, and R. gnavus (Supplemental table 4). For two IPA-predicting species (B. crossotus and F. prausnitzii), higher plasma IPA levels were associated with significantly increased activities of house-keeping pathways involving amino acids biosynthesis, coenzyme biosynthesis, energy metabolism, homeostasis, nucleotide biosynthesis, and nucleotide degradations, while the opposite associations were found in those negative IPA-predicting species (Supplemental figure 2, Panel B).
In the survival analyses, during 218,603 person-years of follow-up, we documented 1,744 T2D cases and 1,129 CHD cases. In the multivariable adjusted model, elevated baseline plasma IPA concentrations were associated with significantly lower T2D risk while higher baseline levels of both dietary tryptophan intake and most plasma tryptophan metabolites from the KP were associated with significantly increased T2D risk (Figure 5). In multivariable-adjusted model, comparing extreme quintiles, the HRs (95% CIs) were 0.70 (0.56, 0.88) for IPA (p trend <0.001), 1.42 (1.20, 1.68) for dietary tryptophan (p trend <0.001), 1.17 (1.02, 1.36) for plasma tryptophan (p trend = 0.02), 1.33 (1.09, 1.62) for kynurenine (p trend <0.01), 1.46 (1.18, 1.79) for quinolinic acid (p trend <0.001), and 1.29 (1.06, 1.56) for xanthurenic (p trend = 0.03). No associations were found for indoxyl sulphate, serotonin, and kynurenic acid (Table 2). In the mediation analysis, KP metabolites and IPA explained 8.2% (2.7%, 22.2%; p=0.003) and 12.2% (4.2%, 30.4%; p=0.01) of the positive association between dietary tryptophan and T2D, respectively, white no statistically significant mediation effects were found for free plasma tryptophan (Supplemental table 5). We also did not find any statistically significant associations with CHD for either IPA or dietary tryptophan and plasma tryptophan metabolites.
Figure 5.
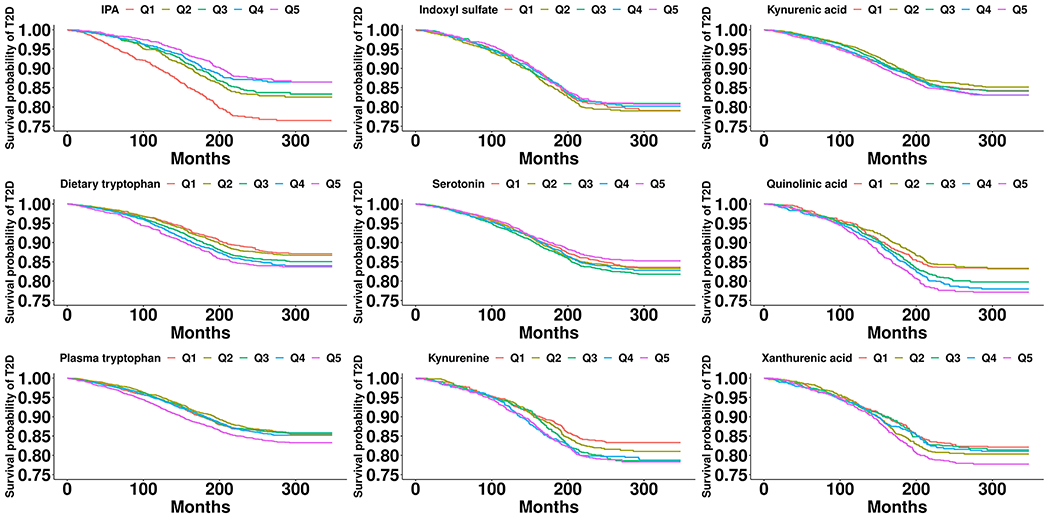
Multivariable-adjusted Kaplan-Meir survival curves for dietary tryptophan and plasma tryptophan metabolites in relation to T2D. The inverse probability weighting was used to adjust for the Kaplan-Meir survival curve. Stabilized weights for each quintile of dietary tryptophan and tryptophan metabolites were calculated using cumulative logistic regression adjusting for age, race, cohort origin, physical activity, alcohol intake, AHEI, smoking status, BMI, and total energy intake.
Table 2.
Associations between dietary tryptophan and tryptophan pathway metabolites and risk of cardiometabolic diseases.
| Baseline dietary tryptophan and plasma tryptophan metabolites | Q1 | Q2 | Q3 | Q4 | Q5 | P trend |
|---|---|---|---|---|---|---|
| Type 2 diabetes | ||||||
| IPA | ||||||
| Case/Person-years | 220/19,155 | 162/20,005 | 153/19,955 | 127/20,571 | 124/21,131 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.84 (0.69,1.04) | 0.86 (0.70,1.06) | 0.68 (0.55,0.85) | 0.70 (0.56,0.88) | <0.001 |
| Indoxyl sulphate | ||||||
| Case/Person-years | 199/21,592 | 218/21,461 | 203/21,721 | 192/21,879 | 196/21,748 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.11 (0.91,1.34) | 0.97 (0.79,1.18) | 1.01 (0.83,1.24) | 0.91 (0.75,1.11) | 0.268 |
| Dietary tryptophan | ||||||
| Case/Person-years | 291/56,064 | 317/55,380 | 364/54,368 | 363/54,831 | 409/52,328 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.05 (0.88,1.25) | 1.27 (1.05,1.54) | 1.29 (1.04,1.59) | 1.50 (1.19,1.90) | <0.001 |
| Plasma tryptophan | ||||||
| Case/Person-years | 348/54,229 | 339/55,766 | 319/55,240 | 349/55,130 | 414/55,080 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.96 (0.82,1.11) | 0.99 (0.85,1.15) | 1.02 (0.88,1.18) | 1.17 (1.02,1.36) | 0.022 |
| Serotonin | ||||||
| Case/Person-years | 335/43,343 | 322/43,110 | 337/42,763 | 294/42,829 | 260/44,114 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.05 (0.90,1.22) | 1.13 (0.97,1.32) | 1.04 (0.89,1.22) | 0.91 (0.78,1.08) | 0.477 |
| Kynurenine | ||||||
| Case/Person-years | 173/22,838 | 168/22,233 | 200/21,740 | 206/21,177 | 261/20,413 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.09 (0.88,1.35) | 1.25 (1.02,1.54) | 1.28 (1.04,1.57) | 1.33 (1.09,1.62) | 0.002 |
| Quinolinic acid | ||||||
| Case/Person-years | 155/22,913 | 161/22,491 | 201/21,943 | 226/21,031 | 265/20,023 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.97 (0.78,1.21) | 1.20 (0.97,1.48) | 1.33 (1.08,1.64) | 1.46 (1.18,1.79) | <0.001 |
| Kynurenic acid | ||||||
| Case/Person-years | 272/47,359 | 281/47,324 | 316/46,572 | 357/46,292 | 376/45,618 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.95 (0.81,1.13) | 1.01 (0.86,1.19) | 1.07 (0.91,1.26) | 1.08 (0.92,1.26) | 0.154 |
| Xanthurenic acid | ||||||
| Case/Person-years | 179/21,650 | 203/21,332 | 159/21,161 | 182/21,055 | 243/20,785 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.11 (0.91,1.36) | 0.99 (0.80,1.22) | 1.05 (0.85,1.30) | 1.29 (1.06,1.56) | 0.032 |
|
| ||||||
| Coronary heart disease | ||||||
| IPA | ||||||
| Case/Person-years | 80/21,454 | 81/21,346 | 69/21,260 | 74/21,511 | 74/21,972 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.04 (0.76,1.42) | 0.97 (0.7,1.34) | 1.06 (0.77,1.46) | 0.96 (0.69,1.32) | 0.843 |
| Indoxyl sulphate | ||||||
| Case/Person-years | 75/23,625 | 94/23,514 | 77/23,761 | 91/23,556 | 83/23,475 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.31 (0.97,1.78) | 1.10 (0.8,1.52) | 1.29 (0.95,1.76) | 1.21 (0.88,1.65) | 0.272 |
| Dietary tryptophan | ||||||
| Case/Person-years | 208/57,633 | 210/57,241 | 239/56,691 | 218/57,422 | 254/55,335 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.01 (0.82,1.24) | 1.05 (0.83,1.32) | 0.89 (0.68,1.15) | 0.88 (0.66,1.18) | 0.295 |
| Plasma tryptophan | ||||||
| Case/Person-years | 247/56,288 | 215/57,850 | 255/56,730 | 223/57,271 | 204/58,884 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.90 (0.75,1.08) | 1.07 (0.9,1.27) | 0.94 (0.79,1.13) | 0.89 (0.74,1.08) | 0.386 |
| Serotonin | ||||||
| Case/Person-years | 129/46,280 | 120/45,958 | 122/45,953 | 109/45,720 | 134/46,180 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.96 (0.75,1.23) | 1.00 (0.78,1.28) | 0.94 (0.73,1.21) | 1.12 (0.88,1.44) | 0.473 |
| Kynurenine | ||||||
| Case/Person-years | 93/24,236 | 75/23,794 | 76/23,644 | 69/23,268 | 107/22,989 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.86 (0.63,1.17) | 0.89 (0.66,1.21) | 0.79 (0.58,1.09) | 1.00 (0.75,1.33) | 0.887 |
| Quinolinic acid | ||||||
| Case/Person-years | 72/24,364 | 79/23,908 | 81/23,739 | 89/23,227 | 99/22,694 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.10 (0.8,1.51) | 1.14 (0.83,1.58) | 1.24 (0.9,1.7) | 1.31 (0.96,1.79) | 0.068 |
| Kynurenic acid | ||||||
| Case/Person-years | 143/49,409 | 107/49,857 | 128/49,255 | 150/49,439 | 149/49,155 | - |
| Multivariable-adjusted model | 1.00(Reference) | 0.69 (0.54,0.89) | 0.83 (0.65,1.06) | 0.98 (0.78,1.23) | 0.94 (0.74,1.18) | 0.612 |
| Xanthurenic acid | ||||||
| Case/Person-years | 93/23,294 | 98/23,135 | 71/22,569 | 64/22,800 | 79/23,407 | - |
| Multivariable-adjusted model | 1.00(Reference) | 1.02 (0.76,1.36) | 1.01 (0.74,1.38) | 0.89 (0.64,1.23) | 0.91 (0.67,1.23) | 0.407 |
Baselines were 1990 in NHS, 1999 in NHSII, and 1994 in HPFS. Proportional hazard model adjusted for age, race, cohort origin, physical activity, alcohol intake, AHEI, smoking status, BMI, and total energy intake.
DISCUSSION
In the current analysis integrating gut microbiome and plasma metabolome, we identified a panel of bacteria species mostly from Firmicutes phylum that were significantly associated with plasma IPA levels. Although the IPA-predicting species did not directly modulate the association between dietary tryptophan intake and plasma IPA, our data showed that such interactions would only manifest with concomitant high dietary fiber intake. These 3-way interactions were persistent not only among major fiber-rich tryptophan food contributors but also observed for subtypes of fiber including soluble fiber, insoluble fiber, and pectin. Using follow-up data from three large prospective cohort studies, a statistically significant inverse association was found between baseline plasma IPA concentrations and incident T2D while higher levels of dietary tryptophan intake and most KP metabolites were associated with significantly increased T2D risk. On the contrary, neither dietary tryptophan nor baseline plasma tryptophan metabolites were associated with risk of CHD.
In comparison with a recent analysis in the Hispanic Community Health Study/Study of Latinos (SOL) study identifying the IPA-predicting microbial genus, we found a quite different panel of taxonomic features at species level which were mostly negative IPA-predicting species.(8) Nevertheless, our results confirmed three Firmicutes genus Eubacterium, Butyrivibrio, and Faecalibacterium as positive IPA-predicting species and the discrepancy might largely reflect the distinct microbial compositions among different ethnicity groups, since this previous study consisted of predominantly Hispanic population as opposed to the mostly non-Hispanic white participants in our study. However, it is important to note that these three identified IPA-associated species are unable to directly produce IPA because they lack the phenyllactate dehydratase gene cluster FldAIBC that is required to encode several key enzymes in IPA biosynthesis.(33) These observations are compatible with the hypothesis that these three positive predictors of IPA and the 14 negative predictors together constitute a microenvironment that is favorable for the IPA-producing species which were not detectable in the current and previous studies. Indeed, our dataset only included one candidate IPA-producing species, Peptostreptococcus stomatis, which were present in less than 2% of the study population, making it challenging to examine its associations with IPA. Notably, none of the IPA-producing species identified from the experimental studies have been successfully identified in the human studies. One probable reason is that not all of these reported IPA-producers have mutualistic relationships with the host and some of the species such as C. botulinum, C. paraputrificum, and P. asaccharolyticus are even detrimental to the host.(34–36) Since our study population was generally healthy without serious cardiometabolic or infectious diseases, the prevalence and abundance of certain pathogenic IPA-producing species were likely to be very low.
To our knowledge, this is the first study among free-living individuals that specifically examined the modulation roles of gut microbiome on the dietary tryptophan-IPA associations. Albeit the null findings in the direct interaction between dietary tryptophan and IPA-associated species on plasma IPA concentrations, we did observe that such interaction was manifested at higher fiber intake level. Consistent with data from the SOL study, fiber intake was not only positively associated with plasma IPA level, but it was also associated with a shared panel of microbial species with IPA.(5,7, and 8) This finding is particularly interesting as no known bacterium can produce IPA from fiber fermentation. Based on the physiological roles of dietary fiber on human health, we propose that fiber may help to activate the tryptophan-gut microbiome interaction via three pathways (Figure 6). First, dietary fiber is known for potentially delaying or decreasing the absorption of macronutrients in the small intestine,(37) resulting in more unabsorbed dietary tryptophan to be metabolized by gut microbiota in colon. Second, higher dietary fiber may inhibit or downregulate the KP in colon and therefore partially shift the tryptophan metabolism towards the indole pathways. This is supported by an experimental study that found butyrate, one of the major fiber fermentation metabolites, could negatively regulate the intestinal epithelial cells indoleamine 2,3-dioxygenase (IDO) expression which is the key enzyme initiating the KP.(38) We also observed consistent inverse associations between fiber intake and kynurenine metabolites in our survival analysis sample (Supplemental table 6). Finally, since the short-chain fatty acids from dietary fiber intake fermentation have profound impacts on bacterial cross-feeding,(39) it may help to shape the microbial communities to benefit the growth or well-being of IPA-producing species, although no study has been conducted to elucidate the cross-feeding networks involving the reported IPA-predictors. Additional studies are clearly warranted to confirm these 3-way interactions as well as the role of dietary fiber in the modulation of tryptophan metabolism in combination with the gut microbiome.
Figure 6.
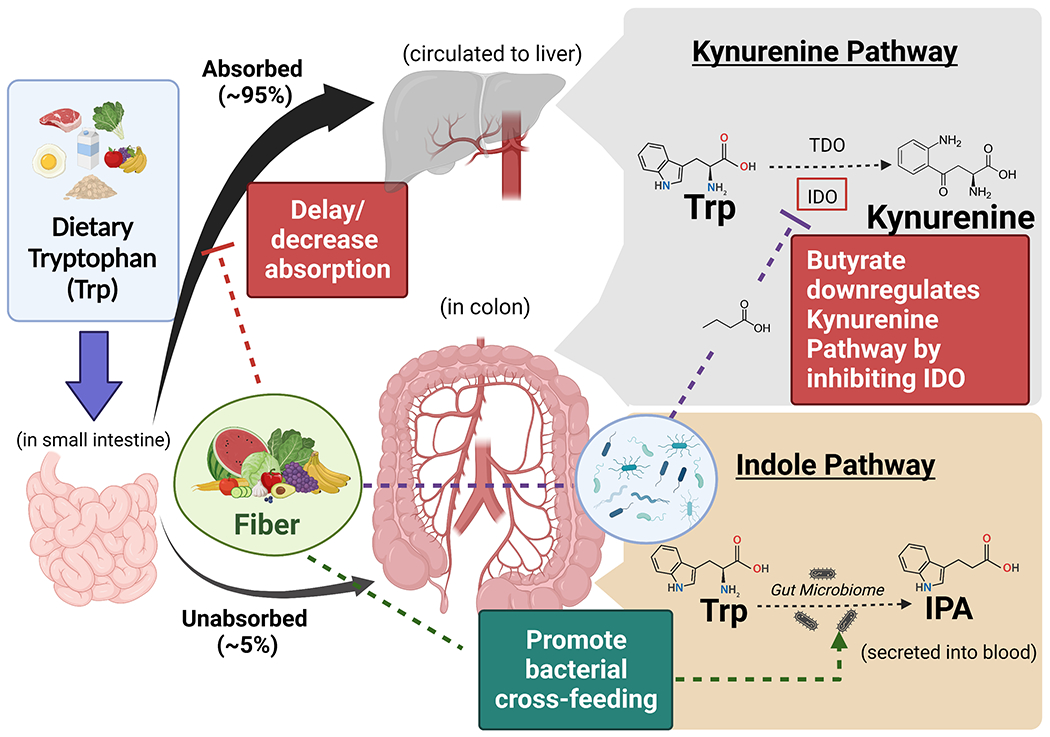
Schematic illustrations of potential roles of dietary fiber modulating the tryptophan metabolism. Three pathways that may shift the tryptophan metabolism towards the indole pathway are proposed: 1) fiber can delay or decrease dietary tryptophan absorption in small intestine, resulting in more unabsorbed tryptophan to be processed in colon; 2) butyrate from fiber fermentation is able to inhibit the key enzyme of the kynurenine pathway in colonic epithelial cells, leaving more tryptophan to be metabolized through the indole pathway; 3) fiber acts as prebiotics that may promote bacterial cross-feeding that particularly benefit the growth or well-being of IPA-producing species.
Findings from the survival analyses showing divergent associations between IPA and KP metabolites in relation to T2D incidence were consistent with previous prospective cohort studies.(4,5,8, and 40) Our study further showed that the positive associations between dietary tryptophan intake and T2D risk were moderately mediated by plasma levels of KP metabolites and IPA. Evidence from both animal and human studies suggest that IPA may be able to prevent T2D development through improving blood glucose, increasing insulin sensitivity, and inhibiting liver lipid synthesis and inflammatory factors.(41,42) Moreover, the similar results found after additionally adjusting for dietary fiber suggested that the inverse associations was likely to be independent of fiber intake.(7) On the contrary, as serum kynurenine/tryptophan ratio has been consistently associated with obesity, metabolic syndrome, and dyslipidemia in population-based studies,(43–45) insulin resistance and inflammation are likely the underlying mechanisms attributed to the positive associations between KP metabolites and T2D risk.(46–48) In addition, although several epidemiological studies have associated KP metabolites with increased risk of CHD and CVD mortality in high-risk or older population,(49–51) our study did not observe statistically significant positive associations. Higher kynurenine/tryptophan ratio resulting from activation of IDO by interferon γ has been suggested as an indicator of increased immunoinflammatory responses in advanced atherosclerosis(52,53) whereas IPA was found to be able to inhibit atherosclerosis by facilitating macrophage reverse cholesterol transport.(54) Given the opposite roles of IPA and KP metabolites implicated in atherosclerosis development and endothelial functions(3,55), further prospective cohort studies are needed to clarify the associations between tryptophan metabolites and cardiovascular health. In addition, data from experiment study and dietary intervention trial are warranted to further interrogate whether fiber and microbiome may jointly modulate the associations between tryptophan intake and diabetes risk.
The major strengths of study included an accurate dietary assessment by diet records, relative comprehensive profiling of metagenomic features that allow for both taxonomic and function analysis, integration of both gut microbiome and plasma metabolome that enabled us to explore the complex relationship among dietary tryptophan, microbial species, fiber, and plasma IPA. Moreover, the survival analyses examining both T2D and CHD provided the prospective data that further substantiated the important pathophysiological roles of tryptophan metabolites on cardiometabolic health. Several limitations also merit discussion. First, our metagenomic functional features were quite limited and did not encompass the key enzymes and pathways in the kynurenine and indole pathways. As a results, the identified enzymes and pathways had mostly house-keeping functions and we were not able to gain additional mechanistic insights in the tryptophan metabolism. Second, the higher order statistical interactions are usually less reliable especially with relatively modest samples size. However, the statistically significant 3-way interactions were not only identified for dietary tryptophan but also among most major tryptophan food contributors across three fiber subtypes, so these results were more likely to reflect plausible biological process than to be merely explained by the chance findings. Third, limited by the modest sample size, we were not able to test any non-linear interactions in the diet-gut microbiome-IPA relationships, which might be more biologically plausible. Fourth, due to the lack of fecal metabolites data, our study could not examine the cross-feeding networks among IPA-associated species which may shed light on the modulation effects of dietary fiber in the tryptophan metabolisms. Finally, our study participants were predominantly white health professionals with higher health consciousness, so whether our findings can be generalized to other ethnic groups or professions needs to be further investigated.
CONCLUSION
In conclusion, the current study identified a panel of IPA-predicting gut microbial species which jointly modulate the associations between dietary tryptophan intake and circulating IPA with dietary fiber in a free-living male population. Plasma IPA was prospectively associated with lower T2D risk while most KP metabolites were associated with increased risk of T2D, but these metabolites were not associated with CHD incidence. Our findings facilitate further understandings of the complex host-microbial crosstalk in tryptophan metabolism and highlight the important role of prebiotics in modulating the relationships in the diet-gut microbiome-disease axis.
Supplementary Material
WHAT IS ALREADY KNOWN ON THIS TOPIC
Among tryptophan catabolites, elevated circulating level of kynurenine pathway metabolites have been associated with increased risk of cardiometabolic disease risk while higher circulating levels of indole pathway metabolites, most notably the indolepropionate (IPA), is associated with a lower risk of type 2 diabetes (T2D).
Different panels of IPA-associated microbial features have been identified in culture-based studies in animal models and free-living human populations.
Dietary fiber as a major prebiotic has been associated with IPA production in a gut microbiota-dependent manner, although how dietary fiber modulates the interactions between dietary tryptophan and gut microbiota profile on circulating IPA concentrations remains to be investigated.
WHAT THIS STUDY ADDS
Utilizing repeatedly measured gut microbiota data and diet records collected among 287 men, we identified 17 microbial species whose abundance was significantly associated with plasma IPA concentrations.
Although IPA-predicting microbial species did not directly modify the associations between dietary tryptophan intake and plasma IPA concentrations, we found that such interactions only manifested among men consuming higher dietary fiber.
A robust inverse association was observed between baseline plasma IPA level and T2D risk while higher plasma concentrations of most kynurenine metabolites were associated with significantly higher T2D incidence. No associations were found in coronary heart disease for any plasma tryptophan metabolites.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings facilitate further understandings of the complex host–microbial crosstalk in tryptophan metabolism and highlight the important role of prebiotics in modulating the relationships in the diet-gut microbiome-disease axis. Additional studies are warranted to further interrogate whether fiber and microbiome may jointly modify the associations between tryptophan intake and T2D risk.
Acknowledgements
We thank the participants and staff of the Nurses’ Health Study, Nurses’ Health Study II, and the Health Professionals Follow-up Study for their valuable contributions. The authors would like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention’s National Program of Cancer Registries (NPCR) and/or the National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) Program. Central registries may also be supported by state agencies, universities, and cancer centers. Participating central cancer registries include the following: Alabama, Alaska, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Hawaii, Idaho, Indiana, Iowa, Kentucky, Louisiana, Massachusetts, Maine, Maryland, Michigan, Mississippi, Montana, Nebraska, Nevada, New Hampshire, New Jersey, New Mexico, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Puerto Rico, Rhode Island, Seattle SEER Registry, South Carolina, Tennessee, Texas, Utah, Virginia, West Virginia, Wyoming.
Funding
The NHS, HPFS, and the current analysis are supported by grants (UM1 CA186107, P01 CA87969, R01 HL034594, U01 CA167552, U01 CA176726, U01 HL145386, R01 HL035464, R01 HL60712, R01 DK120870, R01 DK126698, R01 DK119268, U2C DK129670, DK119268, R01 ES022981, and R21 AG070375) from the National Institutes of Health. The funding sources did not participate in the design or conduct of the study; collection, management, analysis or interpretation of the data; or preparation, review, or approval of the manuscript.
Footnotes
Disclosure statement
All authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Reference
- 1.Taleb S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front Immunol. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oxenkrug G. Insulin Resistance and Dysregulation of Tryptophan–Kynurenine and Kynurenine–Nicotinamide Adenine Dinucleotide Metabolic Pathways. Mol Neurobiol. 2013;48(2):294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song P, Ramprasath T, Wang H, Zou MH. Abnormal kynurenine pathway of tryptophan catabolism in cardiovascular diseases. Cellular and Molecular Life Sciences. 2017;74(16):2899–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Mello VD, Paananen J, Lindström J, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep. 2017;7(1):46337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuomainen M, Lindström J, Lehtonen M, et al. Associations of serum indolepropionic acid, a gut microbiota metabolite, with type 2 diabetes and low-grade inflammation in high-risk individuals. Nutr Diabetes. 2018;8(1):35. doi: 10.1038/s41387-018-0046-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9(1):3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menni C, Hernandez MM, Vital M, Mohney RP, Spector TD, Valdes AM. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes. 2019;10(6):688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Q, Li J, Yu B, et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: an integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut. 2022;71(6):1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences. 2009;106(10):3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang DD, Nguyen LH, Li Y, et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat Med. 2021;27(2):333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulze MB, Manson JE, Ludwig DS, et al. Sugar-sweetened beverages, weight gain, and incidence of type 2 diabetes in young and middle-aged women. JAMA. 2004;292:927–934. [DOI] [PubMed] [Google Scholar]
- 12.Fung TT, Malik V, Rexrode KM, Manson JE, Willett WC, Hu FB. Sweetened beverage consumption and risk of coronary heart disease in women. Am J Clin Nutr. 2009;89(4):1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Ali GS, Mehta RS, Lloyd-Price J, et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol. 2018;3(3):356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta RS, Abu-Ali GS, Drew DA, et al. Stability of the human faecal microbiome in a cohort of adult men. Nat Microbiol. 2018;3(3):347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver LJ, Abu-Ali G, Franzosa EA, et al. bioBakery: a meta’omic analysis environment. Bioinformatics. 2018;34(7):1235–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12(10):902–903. [DOI] [PubMed] [Google Scholar]
- 17.Abubucker S, Segata N, Goll J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8(6):e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paynter NP, Balasubramanian R, Giulianini F, et al. Metabolic Predictors of Incident Coronary Heart Disease in Women. Circulation. 2018;137(8):841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Shaar L, Yuan C, Rosner B, et al. Reproducibility and Validity of a Semiquantitative Food Frequency Questionnaire in Men Assessed by Multiple Methods. Am J Epidemiol. 2021;190(6):1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schakel SF, Sievert YA, Buzzard IM. Sources of data for developing and maintaining a nutrient database1. J Am Diet Assoc. 1988;88(10):1268–1271. [PubMed] [Google Scholar]
- 21.Yuan C, Spiegelman D, Rimm EB, et al. Validity of a Dietary Questionnaire Assessed by Comparison With Multiple Weighed Dietary Records or 24-Hour Recalls. Am J Epidemiol. 2017;185(7):570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Diabetes Data Group. Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes. 1979;28(12):1039–1057. [DOI] [PubMed] [Google Scholar]
- 23.American Diabetes Association. Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20(suppl 1):1183–1197. doi: 10.2337/diacare.26.2007.S5 [DOI] [PubMed] [Google Scholar]
- 24.Manson JE, Stampfer MJ, Colditz GA, et al. Physical activity and incidence of non-insulin-dependent diabetes mellitus in women. The Lancet. 1991;338(8770):774–778. [DOI] [PubMed] [Google Scholar]
- 25.Hu FB, Leitzmann MF, Stampfer MJ, Colditz G a, Willett WC, Rimm EB. Physical activity and television watching in relation to risk for type 2 diabetes mellitus in men. Arch Intern Med. 2001;161(12):1542–1548. [DOI] [PubMed] [Google Scholar]
- 26.Rose G HB Cardiovascular Survey Methods. WHO Monograph Series No. 58. Geneva: World Health Organization.; 1982. [PubMed] [Google Scholar]
- 27.Curb JD, Mctiernan A, Heckbert SR, et al. Outcomes ascertainment and adjudication methods in the women’s health initiative. Ann Epidemiol. 2003;13(9 SUPPL.). [DOI] [PubMed] [Google Scholar]
- 28.Chiuve SE, Fung TT, Rimm EB, et al. Alternative Dietary Indices Both Strongly Predict Risk of Chronic Disease. Journal of Nutrition. 2012;142(C):1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Li Y, Ivey KL, et al. Interplay between diet and gut microbiome, and circulating concentrations of trimethylamine N-oxide: findings from a longitudinal cohort of US men. Gut. 2022;71(4):724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma W, Nguyen LH, Song M, et al. Dietary fiber intake, the gut microbiome, and chronic systemic inflammation in a cohort of adult men. Genome Med. 2021;13(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole SR, Hernán MA. Adjusted survival curves with inverse probability weights. Comput Methods Programs Biomed. 2004;75(1):45–49. [DOI] [PubMed] [Google Scholar]
- 32.Lin DY, Fleming TR, De Gruttola V. Estimating the proportion of treatment effect explained by a surrogate marker. Stat Med. 1997;16(13):1515–1527. doi: [DOI] [PubMed] [Google Scholar]
- 33.Dodd D, Spitzer MH, Van Treuren W, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551(7682):648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa C, Santiago M, Ferreira J, et al. Septic arthritis caused by Peptostreptococcus asaccharolyticus. Acta Reumatol Port. 2016;41(3):271–272 [PubMed] [Google Scholar]
- 35.Shinha T, Hadi C. Clostridium paraputrificum Bacteremia Associated with Colonic Necrosis in a Patient with AIDS. Case Rep Infect Dis. 2015;2015:312919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peck MW. Biology and genomic analysis of Clostridium botulinum. Adv Microb Physiol. 2009;55:183–265,320. [DOI] [PubMed] [Google Scholar]
- 37.Lattimer JM, Haub MD. Effects of Dietary Fiber and Its Components on Metabolic Health. Nutrients. 2010;2(12):1266–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Gallausiaux C, Larraufie P, Jarry A, et al. Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rebnord EW, Strand E, Midttun Ø, et al. The kynurenine:tryptophan ratio as a predictor of incident type 2 diabetes mellitus in individuals with coronary artery disease. Diabetologia. 2017;60(9):1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konopelski P, Mogilnicka I. Biological Effects of Indole-3-Propionic Acid, a Gut Microbiota-Derived Metabolite, and Its Precursor Tryptophan in Mammals’ Health and Disease. Int J Mol Sci. 2022;23(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Jiang M, Zhao J, Song Y, Du W, Shi J. The Mechanism Underlying the Influence of Indole-3-Propionic Acid: A Relevance to Metabolic Disorders. Front Endocrinol (Lausanne). 2022;13:841703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangge H, Summers KL, Meinitzer A, et al. Obesity-related dysregulation of the Tryptophan-Kynurenine metabolism: Role of age and parameters of the metabolic syndrome. Obesity. 2014;22(1):195–201. [DOI] [PubMed] [Google Scholar]
- 44.Mallmann NH, Lima ES, Lalwani P. Dysregulation of Tryptophan Catabolism in Metabolic Syndrome. Metab Syndr Relat Disord. 2018;16(3):135–142. [DOI] [PubMed] [Google Scholar]
- 45.Cussotto S, Delgado I, Anesi A, et al. Tryptophan Metabolic Pathways Are Altered in Obesity and Are Associated With Systemic Inflammation. Front Immunol. 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Bailbé D, Raynal S, et al. Kynurenine-3-monooxygenase expression is activated in the pancreatic endocrine cells by diabetes and its blockade improves glucose-stimulated insulin secretion. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2022;1868(11):166509. [DOI] [PubMed] [Google Scholar]
- 47.Reginaldo C, Jacques P, Scott T, Oxenkrug G, Selhub J, Paul L. Xanthurenic acid is associated with higher insulin resistance and higher odds of diabetes. The FASEB Journal. 2015;29(S1):919.20. [Google Scholar]
- 48.Oxenkrug GF. Role of Kynurenine Pathway in Insulin Resistance: Toward Kynurenine Hypothesis of Insulin Resistance and Diabetes BT - Targeting the Broadly Pathogenic Kynurenine Pathway. In: Mittal S, ed. Springer International Publishing; 2015:169–178. [Google Scholar]
- 49.Pedersen ER, Midttun Ø, Ueland PM, et al. Systemic Markers of Interferon-γ–Mediated Immune Activation and Long-Term Prognosis in Patients With Stable Coronary Artery Disease. Arterioscler Thromb Vasc Biol. 2011;31(3):698–704 [DOI] [PubMed] [Google Scholar]
- 50.Sulo G, Vollset SE, Nygård O, et al. Neopterin and kynurenine–tryptophan ratio as predictors of coronary events in older adults, the Hordaland Health Study. Int J Cardiol. 2013;168(2):1435–1440. [DOI] [PubMed] [Google Scholar]
- 51.Pedersen ER, Svingen GFT, Schartum-Hansen H, et al. Urinary excretion of kynurenine and tryptophan, cardiovascular events, and mortality after elective coronary angiography. Eur Heart J. 2013;34(34):2689–2696. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Liu H, McKenzie G, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16(3):279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niinisalo P, Oksala N, Levula M, et al. Activation of indoleamine 2,3-dioxygenase-induced tryptophan degradation in advanced atherosclerotic plaques: Tampere Vascular Study. Ann Med. 2010;42(1):55–63. [DOI] [PubMed] [Google Scholar]
- 54.Xue H, Chen X, Yu C, et al. Gut Microbially Produced Indole-3-Propionic Acid Inhibits Atherosclerosis by Promoting Reverse Cholesterol Transport and Its Deficiency Is Causally Related to Atherosclerotic Cardiovascular Disease. Circ Res. 2022;131(5):404–420. doi: 10.1161/CIRCRESAHA.122.321253 [DOI] [PubMed] [Google Scholar]
- 55.Cason CA, Dolan KT, Sharma G, et al. Plasma microbiome-modulated indole-and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes. J Vasc Surg. 2018;68(5):1552–1562.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


