Figure 2. Occupancy of mRNA biogenesis and export-related RBPs in Cbp80-containing mRNPs.
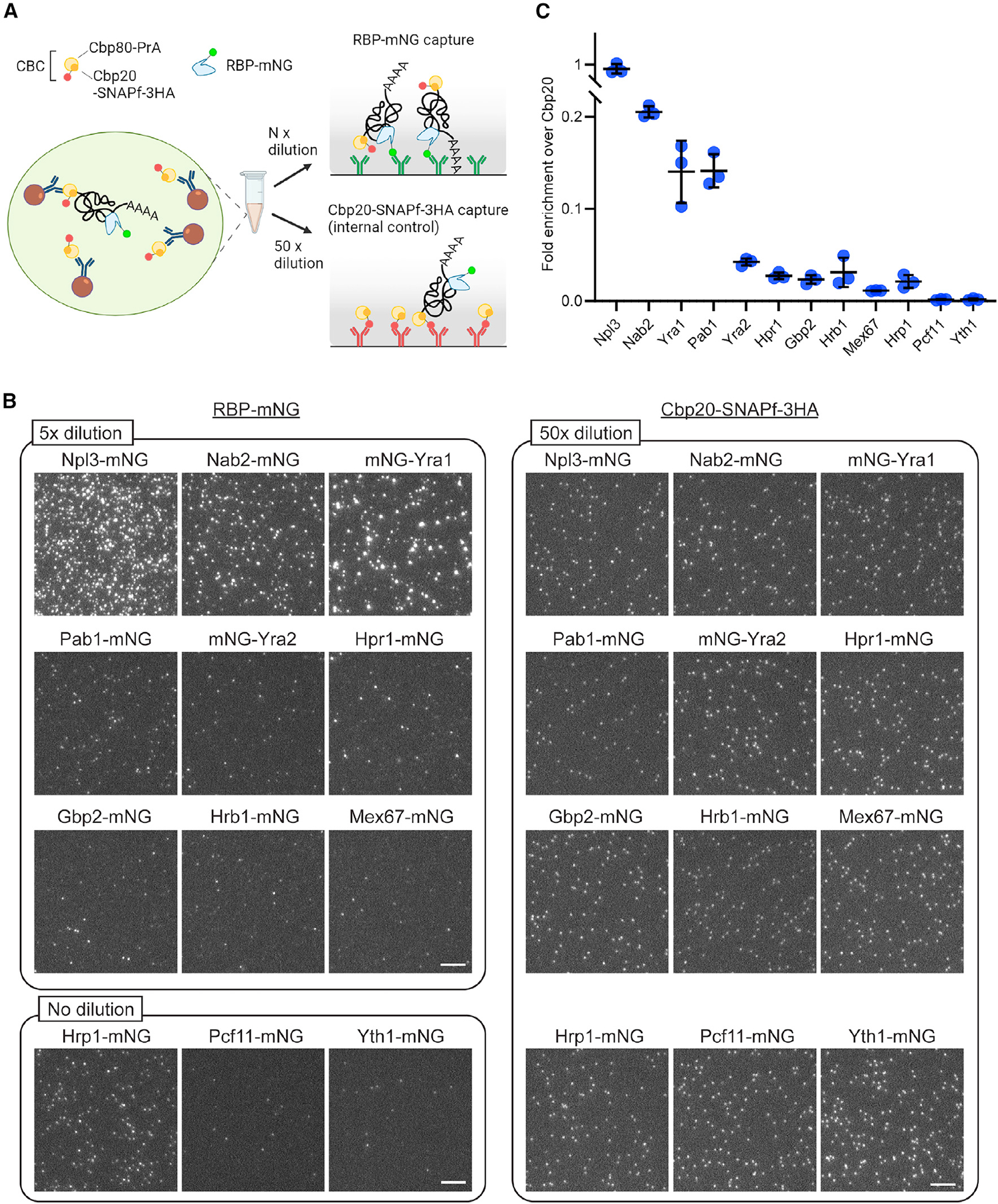
(A) Cartoon summarizes the procedure to analyze the frequency of target RBPs in the population of CBC-containing mRNPs by mRNP-SiMPull. To perform these assays, RBPs were tagged with mNG in a strain with Cbp20-SNAP-3HA with/without Cbp80-PrA. IgG pull-downs were performed to enrich Cbp80-bound mRNPs and CBC itself. The elute was separately diluted and loaded into the mNG antibody coated (to capture RBP-mNG-bound mRNPs) and HA antibody coated (to capture Cbp20-SNAPf-3HA-bound mRNPs and free CBC complexes) slides. The spot number of RBP-mNG counts normalized by Cbp20-SNAPf-3HA counts was used for the comparison between the different RBPs.
(B) Representative images used to determine the frequency of target RBP-containing mRNPs in the population of total Cbp80-bound mRNPs. See Figure S3 for comparison with untagged Cbp80 control strains. Scale bar, 5 μm.
(C) Graph showing the spot number of RBP-mNG normalized by Cbp20-SNAPf-3HA value in the same sample. Mean and standard deviation are shown with individual data points from triplicate experiments. It was noted that mNG tagging of Yra1 and Pab1 caused a growth defect that was tag specific (Figure S3A), but these strains were used in this experiment to maintain consistency.
