Figure 1: Generation of Foxi3GFP and Foxi3CreER mice.
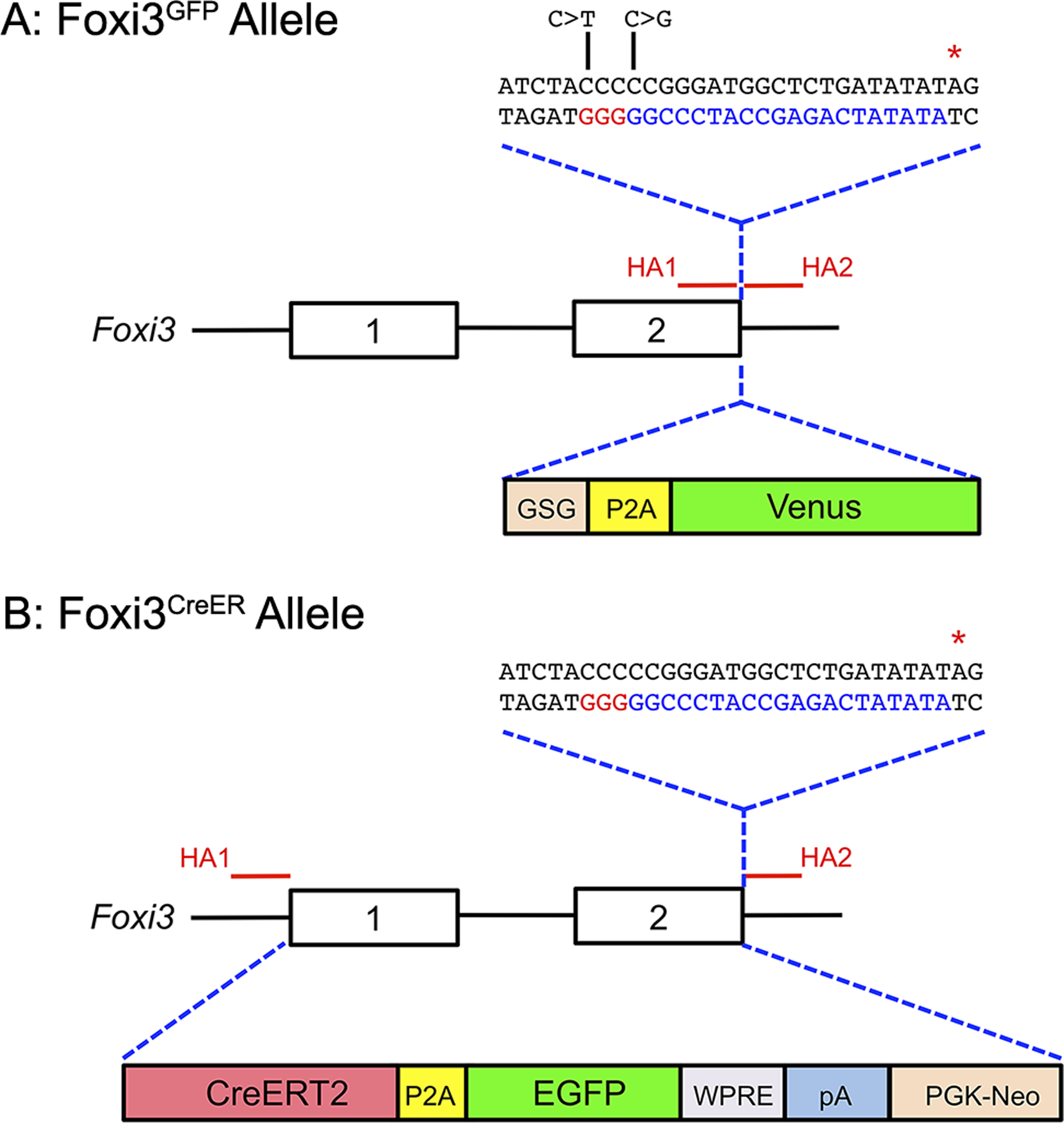
(A) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target the locus by CRISPR-mediated homology-directed repair. The homology arms enclosed a DNA sequence coding for a GSG-P2A-Venus fluorescent protein, followed by a terminating stop codon (red asterisk). The sequence corresponding to the gRNA targeting the 3’ end of Foxi3 is shown in blue letters, with the PAM sequence shown in red letters. The DNA used for homologous recombination contained two nucleotide changes (C>T and C>G; indicated in the diagram) to prevent the inserted sequence from being targeted by the Foxi3 gRNA.
(B) Diagram showing the Foxi3 locus, including the position of the two homology arms (HA1 and HA2) used to target either end of the Foxi3 gene and insert CreER by homologous recombination in ES cells. Exons 1 and 2 and the intervening intron were replaced by an insert containing a CreERT2 fusion cDNA, a P2A sequence followed by EGFP, a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE), a polyA signal, and a PGK-neo resistance cassette. Homologous recombination in ES cells was enhanced by CRISPR-mediated cleavage of the 3’ end of the Foxi3 locus using the same gRNA as in (A) above.
