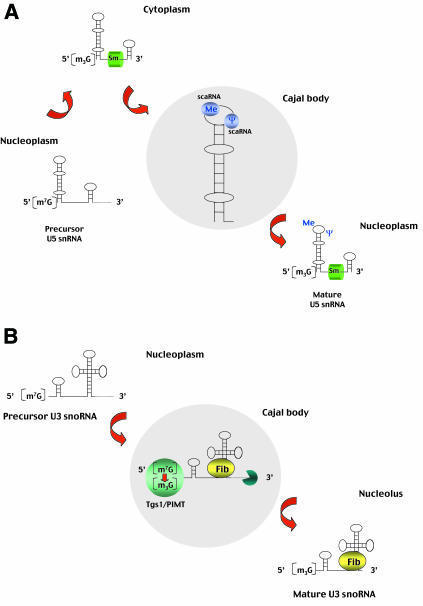
Fig. 1. (A) Processing of spliceosomal U snRNAs. The U1, U2, U4 and U5 spliceosomal snRNAs are synthesized in the nucleoplasm by RNA polymerase II as immature precursors containing a 5′-terminal 7-mono-methylguanosine cap and extra nucleotides at the 3′ end (for simplicity, only the U5 snRNA is depicted). These RNAs are rapidly exported to the cytoplasm, where they bind to Sm proteins. The association of Sm proteins allows subsequent hyper-methylation of the 5′-cap and 3′-end maturation of the snRNA. The newly assembled snRNPs (small nuclear ribonucleoprotein particles) are then transported back into the nucleus, where the snRNAs further undergo 2′-O-ribose methylation (Me) and pseudouridine formation (Ψ). The new data reported by Darzacq et al. (2002) suggest that these modifications are guided by the U85, U87, U88, U89, U90, U91 and U92 small RNAs located in the Cajal body (scaRNAs). Modified nucleotides are confined to the functionally important regions of snRNAs. (B) Processing of the U3 snoRNA. The U3 snoRNA is synthesized in the nucleoplasm by RNA polymerase II as a 3′-extended precursor with a mono-methylated 5′ cap. Cap tri-methylation is catalysed by the methyl-transferase Tgs1/PIMT, which localizes to the Cajal body as shown by Verheggen et al. (2002). Following 3′-end processing, which also takes place within the Cajal body, the U3 RNA binds fibrillarin (Fib) and other core snoRNP proteins. The mature U3 snoRNPs leave the Cajal body to accumulate in the nucleolus.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.