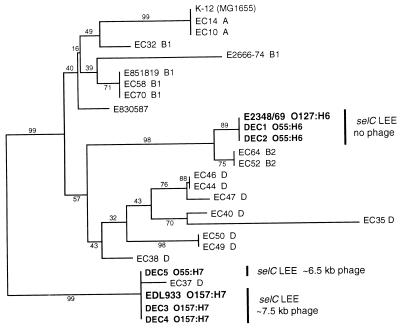
FIG. 5.
Evolutionary relationships among strains used in this study and 20 E. coli reference strains based on mdh sequences. A phylogeny was reconstructed with published data from the ECOR collection (6, 38) (GenBank accession no., U04742 through U04758, U04770, and AF004201) and our EDL933, E2348/69, and DEC strain mdh sequences. The tree is based on a MegAlign (DNASTAR) multiple sequence alignment of positions 34 through 878 of the 916-bp mdh coding region, excluding positions 652 and 653. The phylogeny was reconstructed with a Kimura two-parameter distance matrix and the neighbor-joining algorithim. The percentages of 2,000 bootstrap replicates supporting each cluster are shown along the branches.