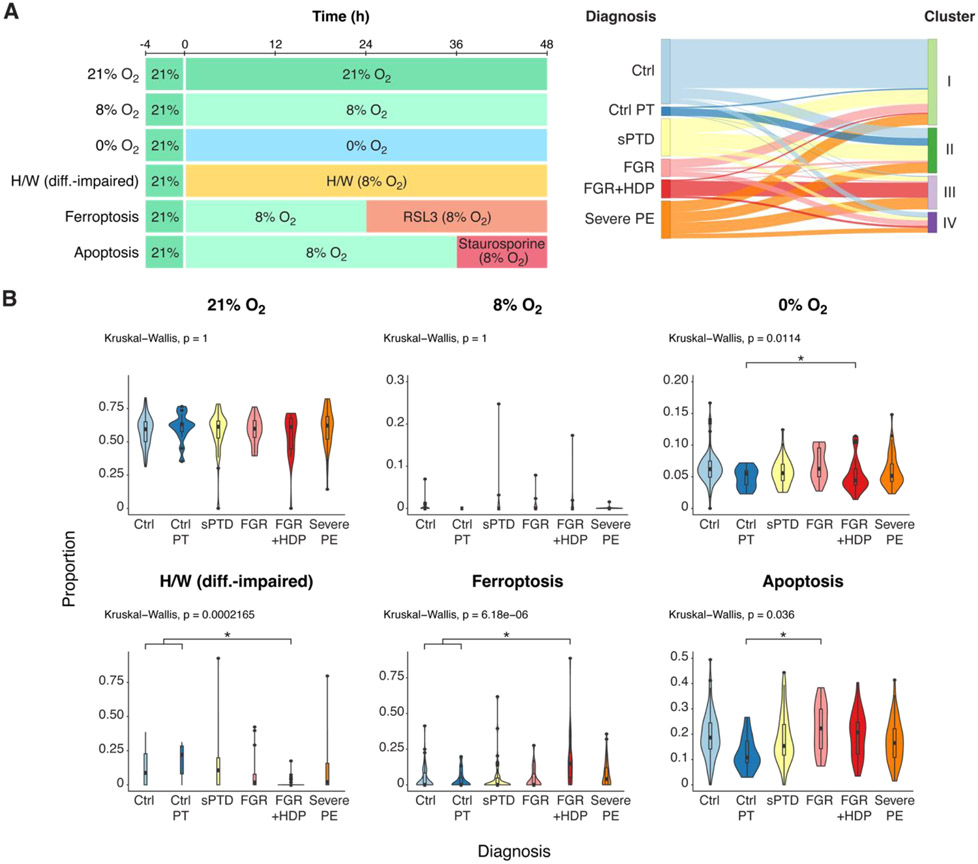
Figure 1. The effect of culture conditions on the representation of PHT cell transcripts among relevant clinical conditions .
(A) An overview of the datasets and experiments used in this study. Left panel: an illustration of the experimental design for PHT cell exposure. PHT cells, isolated from placentas of five uncomplicated term pregnancies, were initially cultured in 20% O2 for 4 h and subsequently exposed to varying oxygen conditions—8% O2, 20% O2, hypoxia at near 0% O2, or to H/W (differentiation-impaired) medium—for 48 h. Cells were exposed to RSL3 induction of ferroptosis (24 h) or staurosporine induction of apoptosis (12 h). At 48 h the cells were harvested for total RNA sequencing. Right panel: a Sankey plot depicting the allocation of placental biopsies from six clinical conditions into four molecular-based clusters (Barak et al, ref. 14), created on the basis of transcriptomic, proteomic, and metabolomic analyses, with Similarity Network Fusion analysis applied to identify clusters (Ctrl, Control; Control-PT, control preterm; sPTD, spontaneous preterm delivery; FGR, fetal growth restriction; FGR+HDP, fetal growth restriction with hypertensive disorder of pregnancy; PE, preeclampsia. (B) A violin plot depicting the effect of culture conditions on the representation of PHT cell transcripts among relevant clinical conditions. The x-axis defines the clinical diagnosis, and the y-axis delineates the proportion of transcripts defining each condition. P-values were calculated by the nonparametric Kruskal-Wallis test, with FDR controlling for multiple comparisons using the Benjamini-Hochberg method. For significant variables the Dunn’s post hoc test was performed. Ctrl, control; Control-PT, control preterm; sPTD, spontaneous preterm delivery; FGR, fetal growth restriction; FGR+HDP, fetal growth restriction with hypertensive disorder of pregnancy; PE, preeclampsia.