Figure 6. SUGP1 recruits DHX15 via its ULM domain.
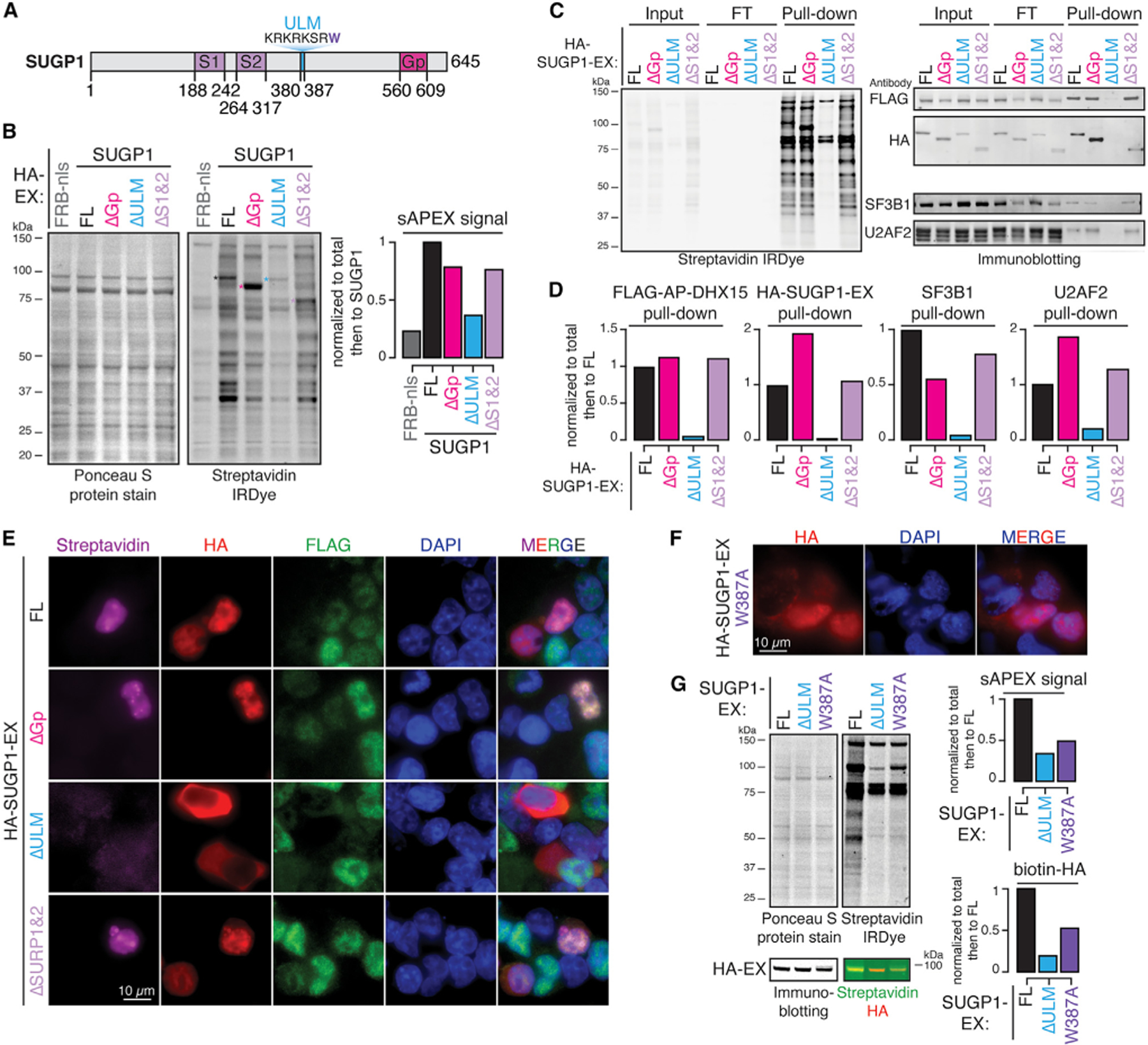
(A) Diagram of SUGP1’s primary domain structure.
(B) Protein blots and quantification of biotinylated proteins labeled by FL versus truncated SUGP1-DHX15 split-APEX activity. Ponceau S protein stain, loading control for total protein; streptavidin IRDye, detection of biotinylated proteins; FRB-nls, FRB control protein fused with an SV40 NLS; ΔGp, G-patch domain truncation; ΔULM, ULM truncation; ΔS1&2, SURP1 and SURP2 truncation; colored asterisks, corresponding bands to FL and truncated HA-SUGP1-EX.
(C) Protein blots of biotinylated proteins labeled in (B) enriched by streptavidin pull-down experiments.
(D) Quantification of pull-downs in (C).
(E) Fluorescent microscopy images of biotinylation signals (streptavidin, magenta), antibodies detecting HA-tagged SUGP1-EX, FL versus truncations (HA, red), FLAG-tagged AP-DHX15 (FLAG, green), nuclear DNA dye (DAPI, blue), and merged channels.
(F) Fluorescent microscopy images of antibody detecting HA-tagged SUGP1.W387A-EX (HA, red), nuclear DNA dye (DAPI, blue), and merged channels.
(G) Protein blots and quantification of biotinylated proteins labeled by full-length (FL), ΔULM, versus W387A mutant SUGP1-DHX15 interaction-reconstituted split-APEX activity. Ponceau S protein stain, loading control for total protein; streptavidin IRDye, detection of biotinylated proteins. Biotinylated HA was detected by merging the streptavidin channel (green) with the HA channel (red). See also Figure S4.
