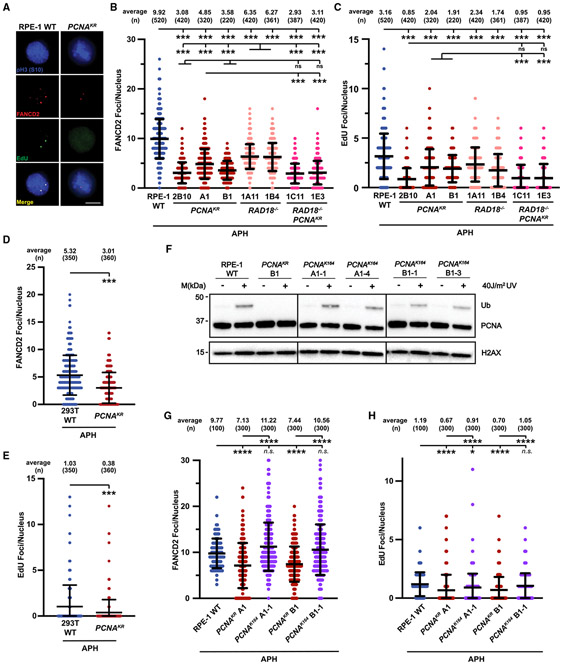
Figure 4. FANCD2-dependent MiDAS is impaired when PCNA K164 cannot be ubiquitinated.
(A) Representative images of phospho-H3 S10-stained nuclei (blue), EdU foci (green), and FANCD2 (red) foci from APH-treated RPE-1 wild-type and PCNAK164R cell lines. EdU foci almost exclusively (>95%) colocalized with FANCD2 foci. Scale bar, 5 μm.
(B) FANCD2 foci quantification from two biological replicates in RPE-1 wild-type (blue), PCNAK164R (maroon), RAD18−/− (salmon), and RAD18−/−;PCNAK164R (pink) cells treated with 300 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by Kruskal-Wallis with Dunn’s multiple comparison test with ***p < 0.001. FANCD2 foci overlapped with EdU foci in 98% of nuclei.
(C) EdU foci quantification from two biological replicates in RPE-1 wild-type (blue), PCNAK164R (maroon), RAD18−/− (salmon), and RAD18−/−;PCNAK164R (pink) cells treated with 300 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by Kruskal-Wallis with Dunn’s multiple comparison test with ***p < 0.001.
(D) FANCD2 foci quantification from two biological replicates in 293T wild-type (blue) and PCNAK164R cells (maroon) treated with 450 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by one-way ANOVA with Tukey’s multiple comparison test with ***p < 0.001.
(E) EdU foci quantification from two biological replicates in 293T wild-type (blue) and PCNAK164R cells (maroon) treated with 450 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by one-way ANOVA with Tukey’s multiple comparison test with ***p < 0.001.
(F) Western blot analyses of chromatin associated PCNA, with or without 40 J/m2 UV treatment, with histone H2AX as the loading control from RPE-1 wildtype, PCNAK164R, and reverted PCNAK164 cells.
(G) FANCD2 foci quantification from two biological replicates in RPE-1 wild-type (blue), PCNAK164R (maroon), and reverted PCNAK164 (purple) cells treated with 300 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by Kruskal-Wallis with Dunn’s multiple comparison test with ****p < 0.001.
(H) EdU foci quantification from two biological replicates in RPE-1 wild-type (blue), PCNAK164R (maroon), and reverted PCNAK164 (purple) cells treated with 300 nM APH. Number (n) of nuclei quantified is listed. Significance was calculated by Kruskal-Wallis with Dunn’s multiple comparison test with ****p < 0.001.