Figure 3. HMCES-DPC self-reversal is dependent on E127 and is stimulated by duplex DNA.
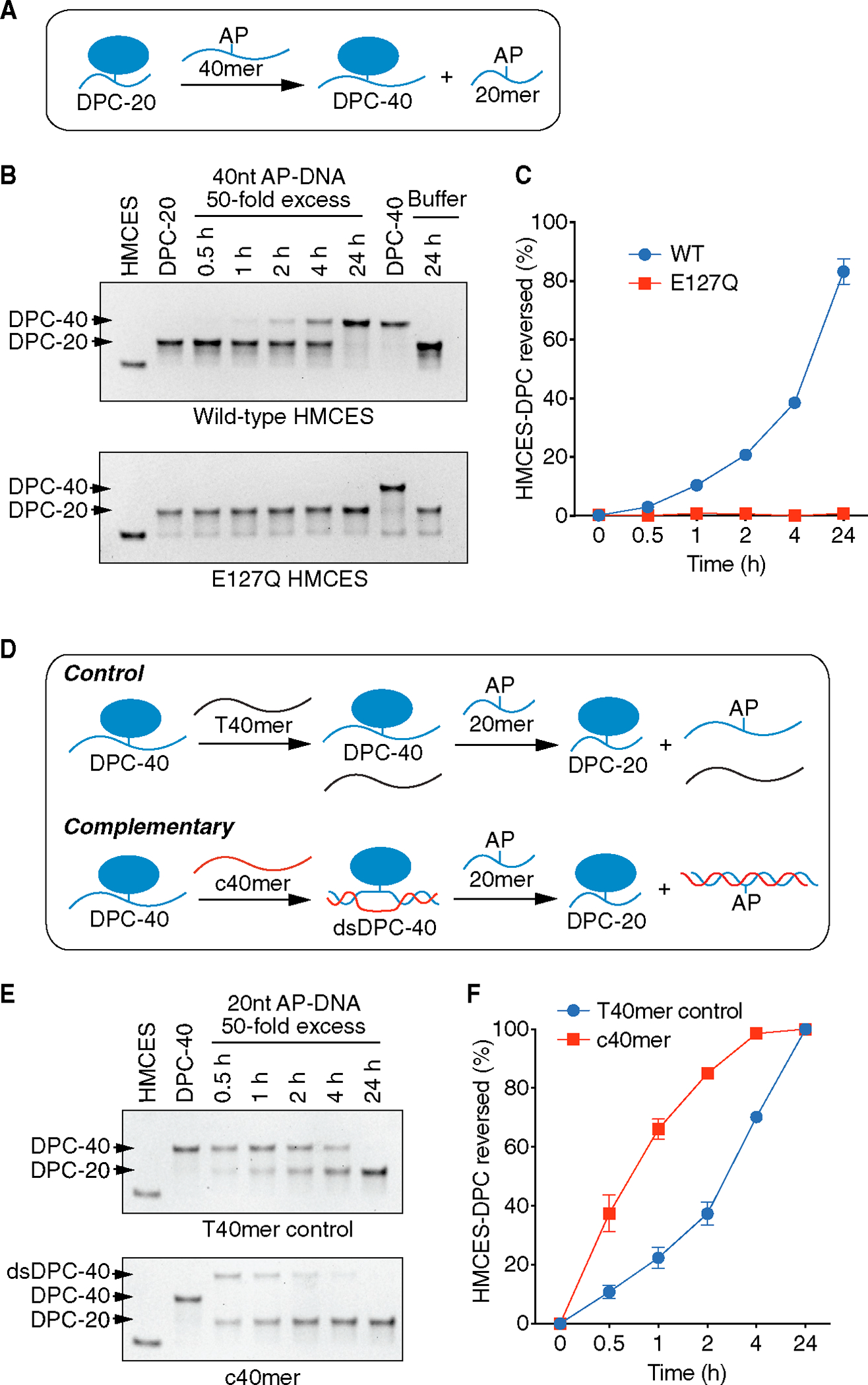
(A) Schematic of HMCES-DPC biochemical self-reversal assay. The AP site containing 40-nucleotide ssDNA oligonucleotide (40-mer) was added at 50× excess.
(B) Representative time course experiment of HMCES-DPC reversal with purified WT or E127Q HMCES. The first lane is HMCES protein without any DNA. The free protein and DPC-20 (DPC with 20-nucleotide oligonucleotide) or DPC-40 (DPC with 40-nucleotide oligonucleotide) were visualized by Coomassie-stained SDS-PAGE.
(C) Quantification of HMCES-DPC reversal. Percent reversed is the amount of DPC-40 product compared to the total. Mean ± SEM, n = 3.
(D) Schematic of HMCES-DPC reversal assay comparing ssDNA vs. duplex DNA. The HMCES DPC-40 was incubated with a complementary (c40mer) or non-complementary (T40mer control) oligonucleotide with a 20-nucleotide ssDNA trap containing an AP site.
(E) Representative time course of HMCES-DPC reversal. The higher migrating band is the product of the hybridization of c40mer with ssDNA HMCES-DPC (dsDPC-40).
(F) Quantification of HMCES-DPC reversal from ssDNA and dsDNA. Percent reversed is the amount of the DPC-20 product compared to the total. Mean ± SEM, n = 3.
