Figure 1.
hPSC-derived DA neurons are susceptible and permissive to SARS-CoV-2 infection
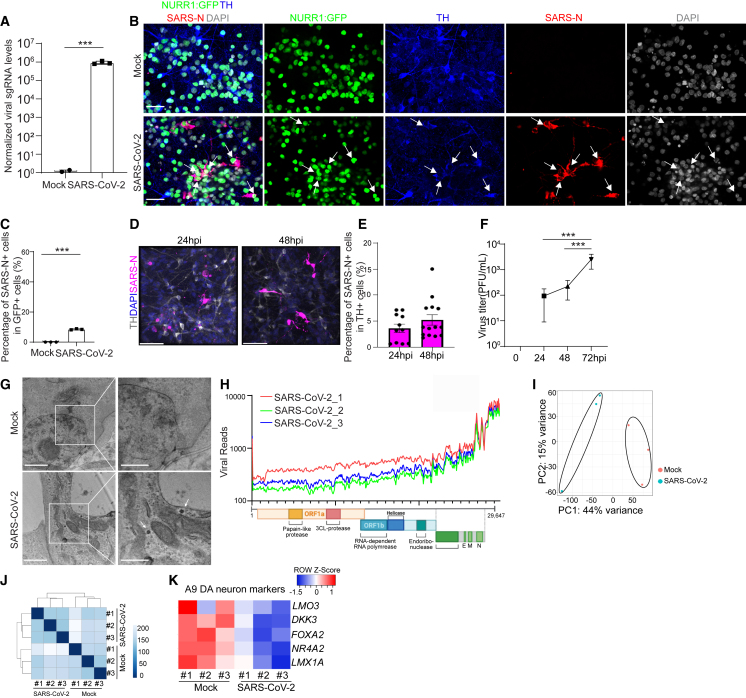
(A) Real-time quantitative PCR analysis of total RNA from purified NURR1:GFP H9-derived DA neurons at 48 hpi of SARS-CoV-2 infection (MOI = 0.2) for viral N sgRNA (small guide RNA). The graph depicts the mean sgRNA level normalized to ACTB. n = 3 independent biological replicates.
(B and C) Representative confocal images (B) and quantification (C) of purified NURR1:GFP H9-DA neurons infected with SARS-CoV-2 (MOI = 0.1) at 72 hpi using antibodies against SARS-CoV-2 nucleocapsid protein (SARS-N) and markers for DA neurons. Scale bars, 50 μm. n = 3 independent biological replicates.
(D and E) Immunostaining (D) and quantification (E) of SARS-N in SARS-CoV-2 infected purified NURR1:GFP H9-derived DA neurons at 24 or 48 hpi (MOI = 0.2). n = 3 independent biological replicates.
(F) Virus endpoint titration assay from supernatants of purified NURR1:GFP H9-DA neurons infected with SARS-CoV-2 (MOI = 0.2) at different time points. n = 3 independent biological replicates.
(G) Transmission electron microscopy (TEM) images of purified NURR1:GFP H9-DA neurons at 72 hpi of SARS-CoV-2 (MOI = 1.0). Arrows point to SARS-CoV-2 viral particles. Right panel: zoom in images. Scale bars, 1 μm. n = 3 independent biological replicates.
(H) RNA-seq read coverage of the viral genome in purified NURR1:GFP H9-DA neurons at 48 hpi (MOI = 0.2). The schematic below depicts the SARS-CoV-2 genome.
(I) PCA plot of gene expression profiles from mock or SARS-CoV-2 infected purified NURR1:GFP H9-DA neurons at 48 hpi (MOI = 0.2).
(J) Clustering analysis of mock or SARS-CoV-2 infected purified NURR1:GFP-DA neurons at 48 hpi (MOI = 0.2).
(K) Heatmap of expression levels of DA neurons and A9 DA neuron marker genes in the mock or SARS-CoV-2 infected purified NURR1:GFP H9-DA neurons at 48 hpi (MOI = 0.2).
Data were presented as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test.
∗∗∗p < 0.001.
See also Figures S1 and S3.