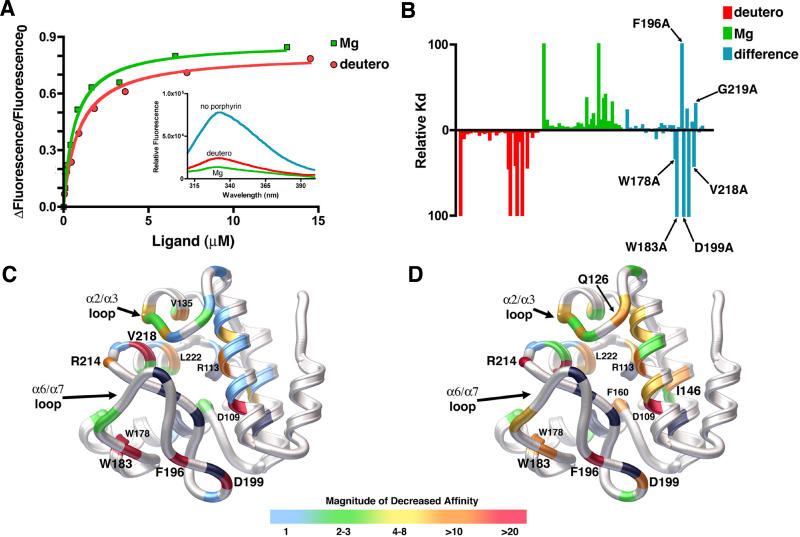
Figure 5. Quantitative Analysis of Porphyrin Binding by SynGUN4.
(A) Comparison of the binding of SynGUN4 to analogs of both Proto and Mg-Proto. Both Mg-Deutero and Deutero quench endogenous tryptophan fluorescence upon binding (inset). A single binding site was assumed for the fitted line.
(B) Relative dissociation constants were determined for each mutant and compared to the wild-type dissociation constant for both Deutero (red bars) and Mg-Deutero (green bars). The difference between these two sets of constants was calculated (blue bars).
(C) Rendered ribbon diagram of the GUN4 core domain with the relative dissociation constants of each mutant for Deutero mapped onto the structure. While in some cases several different amino acid replacements were tested at particular positions, only the results obtained for the alanine mutations are mapped on the backbone structure shown. The x-fold change in the magnitude of the affinity of Deutero for each mutant is color-coded, as depicted by the scale shown at the bottom. Most amino acid changes did not alter binding affinity, as shown by the preponderance of light blue. Of the mutations that measurably alter binding affinity, the majority reside on the “greasy palm” of the GUN4 core domain. Several other energetic hotspots reside on the α2/α3 and α6/α7 loops. Positions of mutations that exhibit a greater than 10-fold decrease in affinity are labeled. Positions colored black failed to produce properly folded protein when mutated to alanine and expressed in E. coli.
(D) Rendered ribbon diagram of the GUN4 core domain with the relative dissociation constants of each mutant for Mg-Deutero mapped onto the structure. Color coding is the same as for (C). In contrast to Deutero binding, many more mutants alter in vitro binding as shown by the lesser amount of light blue and the prominence of green and yellow color coding.