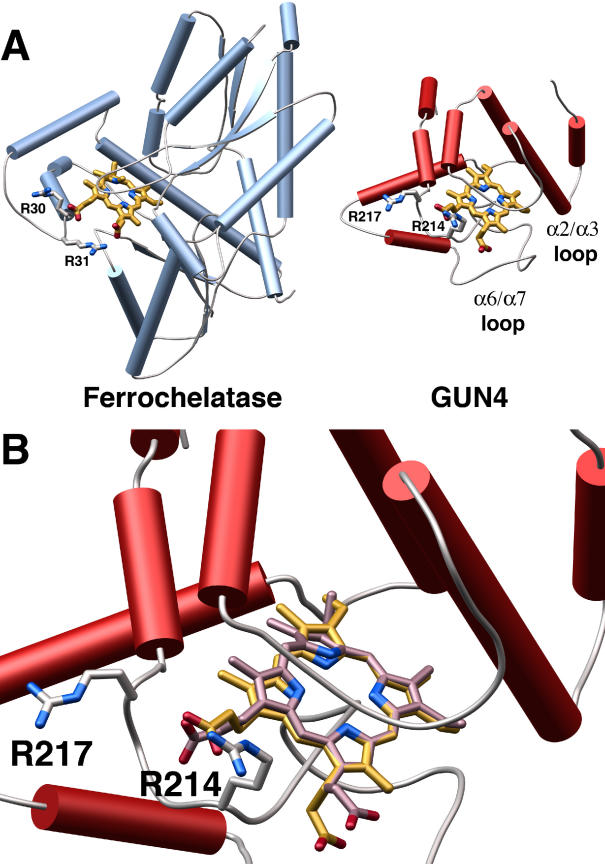
Figure 6. Model of a Putative SynGUN4 Porphyrin Complex Compared to an Experimentally Determined Structure for Ferrochelatase Bound to NMMP.
(A) Comparison of the crystal structure of the B. subtilis ferrochelatase bound to NMMP to the model of the SynGUN4 core domain bound to Mg-Proto. The SynGUN4 core domain • Mg-Proto model was generated by GOLD [54]. The carboxylic acid moieties of the porphyrin were staggered between the δ-guanido side chains of Arg214 and Arg217. The position of the arginine loop used to tether the carboxyl moieties of the porphyrin bound to ferrochelatase served as the fixed point for the structural alignment of SynGUN4 and ferrochelatase.
(B) Close-up view of the structural alignment between Mg-Proto (gold) and NMMP (lavender). Attempts to strictly superimpose all of the atoms of the two porphyrins resulted in at least one corner of the porphyrin scaffold residing out of the plane defined by the flat Mg-Proto complex, because of the pucker of NMMP.