Figure 7. Shared transcriptional changes across DNMT3A mutants indicate disruption of growth and synaptic processes.
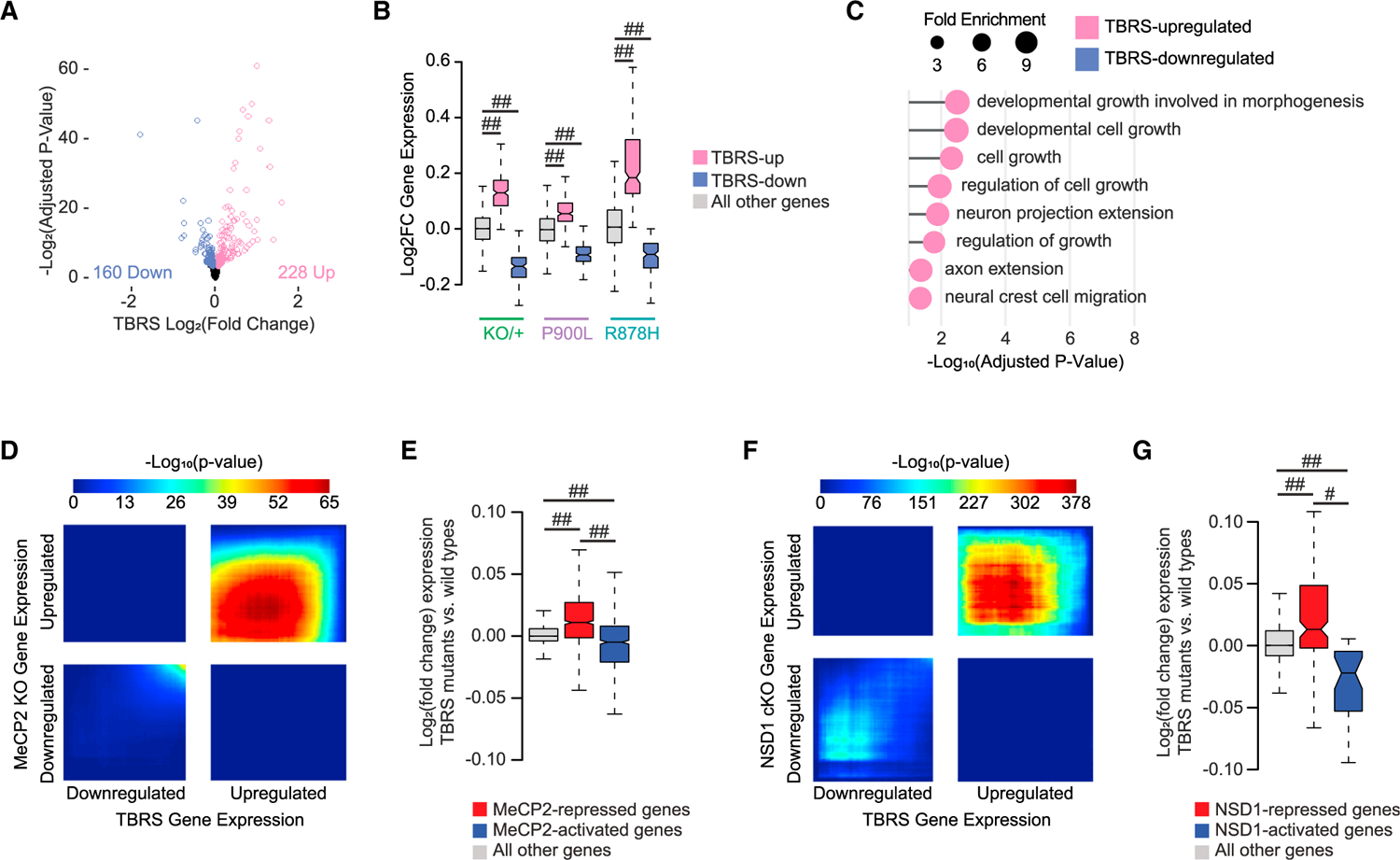
(A) Volcano plot of DESeq2 log2 fold changes from DNMT3A mutant vs. WT analysis between paired littermate data from Dnmt3aKO/+,9 Dnmt3aR878H/+, and Dnmt3aP900L/+ datasets (design = ~pair + group; contrast by group). Genes reaching a significance of false discovery rate (FDR) <0.1 are indicated.
(B) Log2 fold changes of gene expression within each mutant (KO/+, P900L, and R878H) of genes defined as differentially expressed in the combined TBRS mutant analysis.
(C) Most significant PANTHER Gene Ontology biological process terms enriched in the TBRS differentially expressed gene lists. No significant terms were identified in the TBRS-downregulated gene list.
(D) RRHO of transcriptome-wide gene expression changes in the cerebral cortex of TBRS mutants vs. MeCP2 KO mice.20
(E) Log2 fold changes in the TBRS mutants at genes significantly disrupted in MeCP2 mutants.20
(F) RRHO of transcriptome-wide gene expression changes in the cerebral cortex of TBRS mutants vs. NSD1 conditional KO mice.42
(G) Log2 fold changes in the TBRS mutants at genes significantly disrupted in NSD1 conditional KO (cKO) cortices.42
Notched box and whisker plots indicate median, interquartile, and confidence interval of median with significance from Wilcoxon rank-sum test shown. Detailed statistics and sample sizes are in Table S1. #p < 0.0001; ##p < 2e–10.
