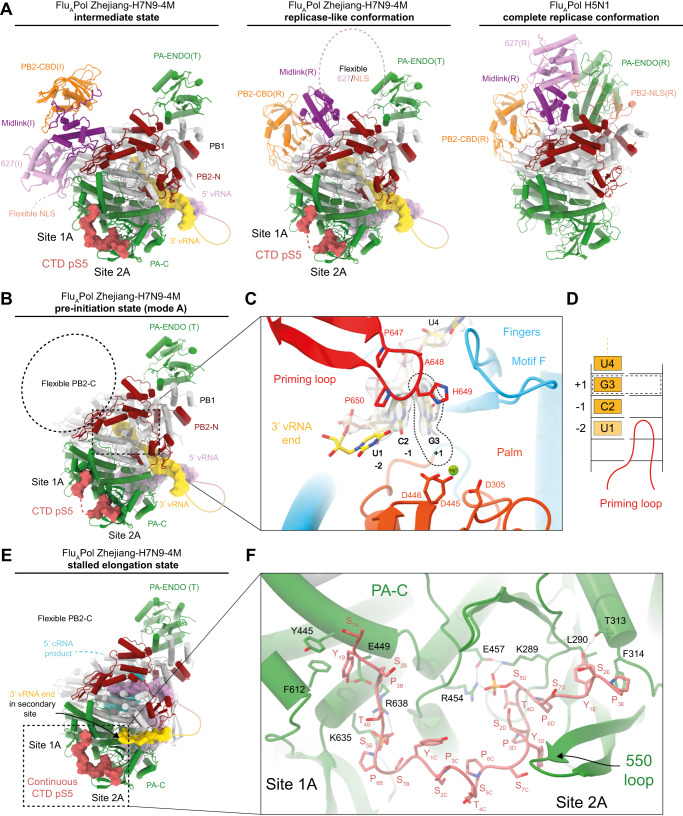
Fig. 4. FluPol replication is consistent with CTD-binding to the FluPol.
A Left: Cartoon representation of FluAPol Zhejiang-H7N9-4M in the intermediate conformation (obtained in this study) as previously described14,45, bound to pS5 CTD. Middle: FluAPol Zhejiang-H7N9-4M in a replicase-like conformation bound to pS5 CTD. Right: FluAPol replicase conformation from A/duck/Fujian/01/2002(H5N1) (PDB: 6QPF)41. FluPols are aligned on the PB1 subunit. FluAPol Zhejiang-H7N9-4M PA-ENDO remains in a transcriptase conformation (PA-ENDO(T)). In FluAPol Fujian-H5N1 structure, it rotates and interacts with the PB2-NLS domain (PA-ENDO(R)). PA is coloured in green, PB1 in light grey, PB2-N in dark red, PB2-CBD in orange, PB2 mid-link in purple, PB2 627 in plum, PB2 NLS in salmon. The pS5 CTD is coloured in red, displayed as surface, with discontinuity between sites 1A/2A shown as a dotted line. PB2-627/NLS domains flexibility is highlighted as a dotted circle. RNAs are displayed as surfaces. The 5′ vRNA end is coloured in pink, the 3′ vRNA end in yellow. Flexible nts are represented as solid line. B Cartoon representation of FluAPol Zhejiang-H7N9-4M structure in the pre-initiation state mode A. Colour code is identical to (A). PB2 C-terminal domains (PB2-C) are flexible, highlighted as a dotted circle. C Close-up view on the 3′ vRNA end in FluAPol Zhejiang-H7N9-4M active site. The Coulomb potential map of the template is shown. 3′-U1 remains unseen in the map. The 3′-G3 is in the +1 active site, highlighted by a dotted line. The priming loop is coloured in red, the palm domain in orange with the catalytic aspartic acids displayed, coordinating a Mg2+ ion, in green. The motif F is coloured in blue. D Schematic representation of the 3′ vRNA end terminal nucleotides active site position, as seen in (C). E Cartoon representation of FluAPol Zhejiang-H7N9-4M in stalled elongation state. The colour code is identical to A. PB2-C domains are flexible. The cRNA de novo replication product is displayed as surface when visible or as a dotted line when flexible. The 3′ vRNA end is bound to the secondary site. F Close-up view on the pS5 CTD interaction with FluAPol Zhejiang-H7N9-4M PA-C domain. FluPol residues interacting with the pS5 CTD are shown. Each CTD pS5 repeat is indicated.