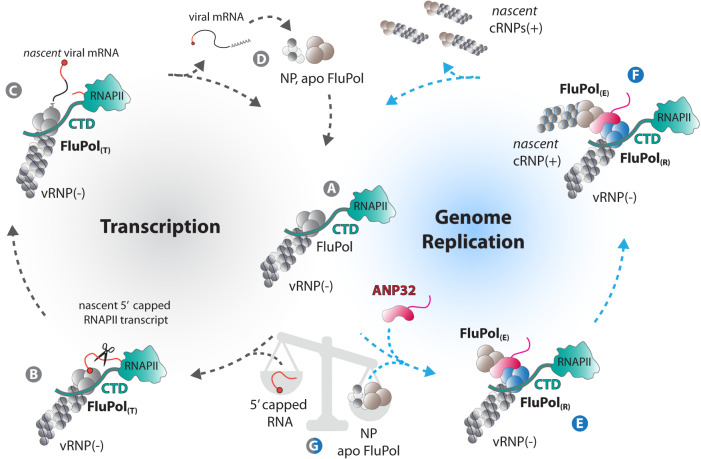
Fig. 6. A model for RNAP II CTD-anchored transcription and replication of the influenza virus genome.
A model for RNAP II CTD-anchored transcription and replication of the influenza virus genome. Upon influenza virus infection, incoming vRNPs are imported into the nucleus and bind to the host RNAP II CTD through bipartite interaction sites on the FluPol (A). This intimate association allows FluPol, in the transcriptase conformation (FluPol(T)), to cleave short capped oligomers derived from nascent RNAP II transcripts in a process referred to as ‘cap-snatching’ to initiate primary transcription of viral mRNAs (B). Polyadenylation is achieved by a non-canonical mechanism involving stuttering of the viral polymerase at a polyadenylation signal (C). The 5′ and 3′ vRNA extremities always remain bound to the polymerase which allows efficient recycling from the termination to the initiation state (C–A). Upon translation of viral mRNAs, de novo synthesised FluPols in an apo state (not viral RNA-bound) and NPs are imported into the nucleus (D). The apo FluPol, in conjunction with the host factor ANP32, associates with the parental CTD-associated FluPol(T) and triggers its conformational transition into a replicating FluPol(R), to form an asymmetric FluPol(R)-FluPol(E) dimer (E) where FluPol(E) is encapsidating the newly synthesised cRNA in conjunction with NP (F). The FluPol(R)-ANP32-FluPol(E) replication complex remains associated to the RNAP II through direct binding of the CTD to FluPol(R). Anchoring of the parental vRNP to the CTD allows it to engage into successive cycles of either viral genome replication or mRNA transcription, depending on the availability of NP, apo FluPol and/or nascent capped oligomers derived from actively transcribing RNAP II (G). Such switching between both activities allows efficient adaptation to waving levels of de novo synthesised vRNP components in the nucleus of an infected cell and is key to ensure a correct balance between genome replication and mRNA transcription.