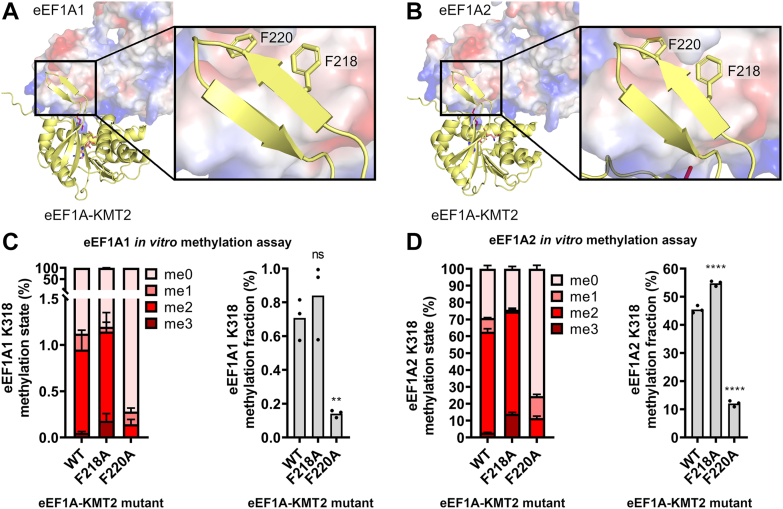
Figure 7.
A conserved phenylalanine in human eEF1A-KMT2 is critical for its eEF1A K318 methylation activity.A and B, AlphaFold-Multimer model showing a beta-hairpin extending from eEF1A-KMT2 (isoform 207) binding a hydrophobic pocket in domain 2 of eEF1A1 (A) or eEF1A2 (B). eEF1A-KMT2-207 (yellow) is shown as a cartoon structure. eEF1A1/2 is shown as its surface electrostatic potential (blue = positive, red = negative, white = neutral). Inset: sidechains of conserved eEF1A-KMT2-207 residues F218 and F220 on its beta-hairpin are shown as sticks. C and D, in vitro methylation assays of eEF1A-KMT2 mutants. Purified WT and mutant eEF1A-KMT-207 (3 μM) were incubated with eEF1A1 (C) or eEF1A2 (D) (2.2 μM) in the presence of AdoMet for 2 h at 37 °C. Proteins were separated by SDS-PAGE (see Fig. S21), eEF1A1/2 gel bands digested with AspN, and the resulting eEF1A1/2 K318 methylation was detected by LC-MS/MS and quantification of AspN-generated peptide DNVGFNVKNVSVK (K318 underlined) in its triply-charged state. Left: Relative levels of eEF1A1/2 K318 methylation states. Right: eEF1A1/2 K318 methylation fraction relative to 100% trimethylated K318. Methylation fractions from mutant eEF1A-KMT2 were compared to WT eEF1A-KMT2 using an ordinary one-way ANOVA with a post hoc Dunnett’s multiple comparisons test (ns: not significant, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001).