FIGURE 1.
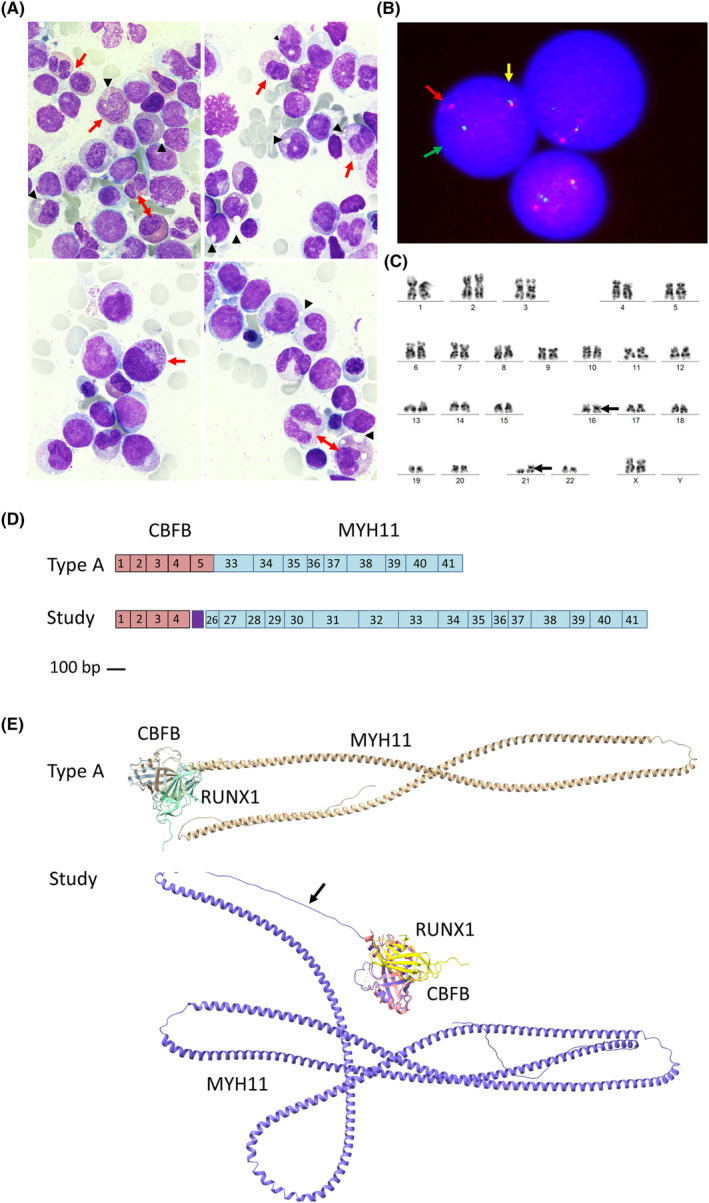
Characterization of atypical inv(16)(p13.1q22), CBFB::MYH11. (A) Bone marrow smear with May Grunwald‐Giemsa (×1000, lower panels are additionally magnified) showing infiltration by myelomonocytic blasts and 15% abnormal eosinophils and their precursors, most of them with abnormal proeosinophilic granules with a characteristic barely pinkish‐orange staining (red arrows). In addition, a remarkable cytoplasmic vacuolization was observed, which was not stained (black arrowheads). (B) FISH of bone marrow cells using the CBFB break apart probe (LSI CBFB Dual Color Break Apart Probe; MetaSystems, Germany) showing one fusion signal (unsplit CBFB, 16q21‐22, yellow arrow), one green signal (split CBFB, 5′ end, green arrow) and one red signal (split CBFB, 3′ end, red arrow). (C) Giemsa‐banded karyotype of bone marrow cells at diagnosis: 46,XX,inv(16)(p13q22),add(21)(p10)[14]/46,XX[6]. (D) Graphical representation of the common type A and the new atypical transcript found in this study type A. Scale representation (scale bar 100pb) of the contribution of the exons of CBFB (red boxes) and MYH11 (blue boxes) to each transcript. The linker peptide of the atypical transcript not belonging to exonic sequences is shown (purple boxes). (E) AlphaFold in silico modelling of the CBFB::MYH11 fusion proteins, and RUNX1 interaction, of the common type A and of the new aberrant inversion found in this study. The linker peptide is indicated with an arrow. Results were visualized with UCSF ChimeraX v1.4.
