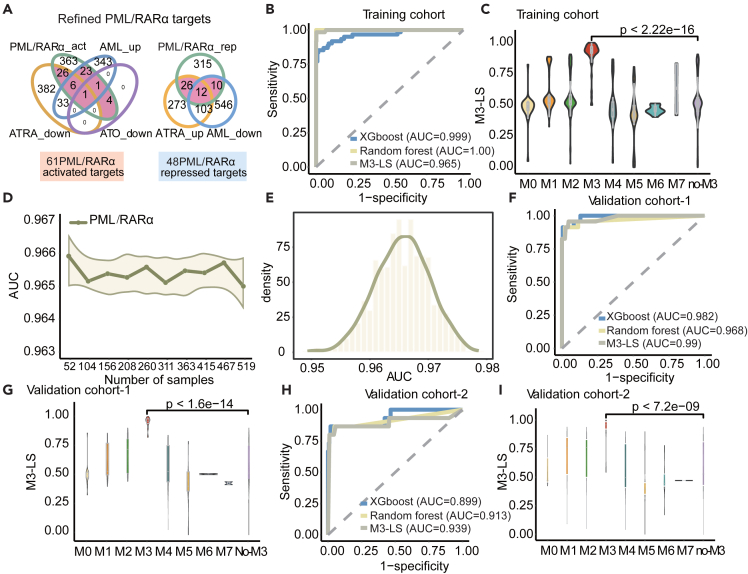
Figure 3.
Performance of M3-LS model was improved by integrating ATO/ATRA response genes
(A) Venn plot of model optimization, including leading edge genes (LEGs) of ATO, differential genes robust rank aggregation results of ATRA, PML/RARα target genes, and differential expression genes of M3 and healthy controls in the training cohort.
(B) Random forest, XGBoost machine learning model and optimized M3-LS index were used to predict M3 samples, and ROC analysis was used to evaluate the prediction model in the training cohort.
(C) Comparison of the scores of the optimized models for each subtype.
(D) The model was validated using ROC analysis. Use the model to predict several randomly selected samples, the line graph represents the size of the AUC.
(E) Probability density distribution plot of AUC.
(F) Random forest, XGBoost machine learning model and optimized M3-LS index were used to predict M3 samples, and ROC analysis was used to evaluate the prediction model in the validation cohort-1.
(G) Comparison of model scores for each subtype in the validation cohort-1.
(H) Random forest, XGBoost machine learning model and optimized M3-LS index were used to predict M3 samples, and ROC analysis was used to evaluate the prediction model in the validation cohort-2.
(I) Comparison of model scores for each subtype in the validation cohort-2. Statistics were calculated using Wilcoxon Rank-Sum test.