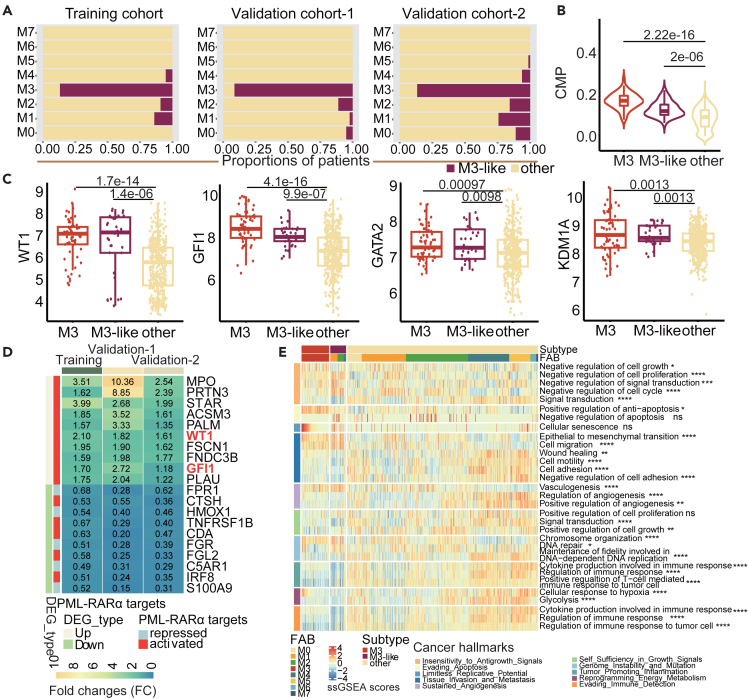
Figure 4.
M3-LS model identifies additional patients like M3 subtype
(A) The proportion of M3-like samples predicted by the optimized model in each subtype in the training and validation cohorts, respectively. Amaranth represents the proportion of samples predicted to be M3-like subtype, and yellow represents the proportion of samples not predicted to be M3-like subtype.
(B) Violin plot of the proportion of common myeloid progenitor (CMP) of each subtype identified in the training cohort. Boxes plots show median, 25th and 75th percentiles of CMP for each subtype. p values are calculated using Kruskal-Wallis Test.
(C) The expression levels of WT1, GFI1, GATA2, and KDM1A of each subtype were compared AML cases with predicted as M3-like versus M3 subtype and other samples in the training cohort. p value was estimated using Kruskal-Wallis Test.
(D) The differential expression of PML/RARα target genes in M3-like and other samples in the training and validation cohorts. The heatmap shows the fold change (FC) values of differential genes in M3-like samples relative to other samples, and the genes in red font are characteristic genes of M3 subtype.
(E) Cancer Hallmark pathway enrichment of M3 subtype, M3-like subtype and other samples. The heatmap shows the results of single sample gene set enrichment analysis (ssGSEA) of each subtype sample in each Cancer Hallmark pathway (Statistical significance was assessed by Wilcoxon Rank-Sum test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Data are represented as mean.