 Cochrane (logo), Covidence (figure)
Cochrane (logo), Covidence (figure)
Since the birth of systematic reviews, technology has been an integral part of efforts to understand health evidence. Nevertheless, review authors commonly conduct the majority of their work on a patchwork of general software products poorly adapted to their needs, much of the data they handle is not captured for future use, and the core review output of a static PDF document limits the ability to search and process the contents of the review.
In recent years a combination of increasing frustration with the limitations of current systematic review technologies, an awareness of the impact technological developments have had in other fields, and promising results of recent innovations have led to an increasing focus on the opportunities afforded by emerging technologies. To help move the field forward Cochrane convened the #CochraneTech Symposium immediately prior to the 21st Cochrane Colloquium in Quebec City, Canada, in 2013. Following the success of this event, the second #CochraneTech Symposium will be held in Hyderabad, India, on Saturday 20th September (tech.cochrane.org/cochranetech).
On the eve of the second #CochraneTech Symposium, we reflect here on the current state of systematic review technology. As Ida Sim asked in her opening plenary at the first #CochraneTech Symposium: how can we better leverage technology and knowledge, not only to help us prepare systematic reviews more efficiently, but also to deliver the outputs better suited to our end‐users?
A diverse set of technologies may be used to produce a Cochrane Review. Ongoing innovation in this area is creating better options for authors. See, for example, these lists of available tools. At last year's Symposium three key goals for these tools were proposed to guide Cochrane's engagement: authors should be able to make easy, informed choices of tools for their Cochrane reviews; authors should have seamless transfer of data between software tools; and Cochrane as an organisation should support tools which make Cochrane Review data reusable.
Additional benefit can be derived from machine processes that assist review authors by semi‐automating key review tasks. The most well‐advanced area is citation screening, where there is some evidence of reduced workload without sacrificing thoroughness. A similar data mining approach might also enable the automated assignment of trials upon publication in major databases (e.g. PubMed and Embase) to individual review groups and then to individual reviews. Similarly, machine assistance with the extraction of data from study publications has been explored in several studies and would make a substantial contribution to the efficiency of review production.
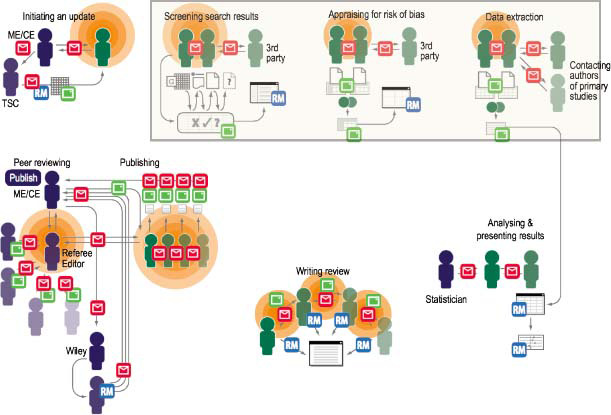
Opportunities for improvement: Cochrane review updating workflow with pain points and communication flows
Despite the global network of contributors to Cochrane, the exponential growth in primary research and increasing methodological expectations is outstripping the capacity of traditional contributor models. Enabling contributions from a much broader contributor base using a crowdsourcing model is being explored in the ongoing Embase project, discussed at last year's Symposium, with an update expected at this year's meeting. A key topic this year will also be the breadth of tasks amenable to a crowdsourced approach, including translations, data extraction, and meta‐data tagging.
To succeed in meeting the needs of end users it is critical that content and delivery be tailored to different audiences and contexts, but it must also be possible for users to combine content from different sources. This flexibility can be achieved by better structuring of Cochrane content, assigning unique identifiers for individual studies, mapping data to ontologies (standard computable ways of describing complex abstractions), and defining descriptors of study settings. A key recommendation from last year's Symposium was for Cochrane to invest in better structuring of content. Since then, substantial work has been undertaken in the development of a PICO ontology and in the broader Cochrane ‘linked data’ project. This work lays the foundation for Cochrane content to move ‘beyond the PDF’ and be more easily navigable, reusable, and traceable.
We will always need to draw the soundest inference from the totality of the evidence, but both the inputs and outputs are changing. Individual patient data from randomised trials, large cohort studies with a million or more participants, health system data, and personal digital technologies will increasingly make ‘big data’ available. As pointed out by last year's second plenary speaker, John Wilbanks, there are opportunities that arise when these data are open. Open systems produce greater value over the long term because they lead to unanticipated change from unanticipated people in unanticipated ways. The challenge will be accommodating diverse forms of evidence of very different levels of rigour and helping users understand and adjust for the limitations of these data.
In describing Cochrane as an intervention, Dr Sim characterised a PICO question with the future intervention still evidence synthesis ‘the Cochrane way’, but reimagined in response to changing circumstances: the comparator likely being big data rather than the current ‘eminence‐based medicine’; the target population for systematic review findings increasingly patients themselves and their families; outcomes increasingly including both population‐level and individual‐level evidence as well as a stronger emphasis on costs; and timescales shifting from years to continuous review updating.
The goal is to combine people, data and computational methods to enable everyone to get the most sound and up to date individual and population level summary evidence for decision making. As Dr Sim stated in closing her plenary presentation, “Cochrane is the only group that can and should take the major lead in much of this work. We need Cochrane now more than ever.”
Acknowledgements
The authors would like to thank John Wilbanks (Sage Bionetworks) and all the participants at the 2013 #CochraneTech Symposium for their contributions.
Feedback on this editorial and proposals for future editorials are welcome.
End Notes
Declarations of interest
The authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available upon request). JE is co‐founder of Covidence, a non‐profit web platform for systematic review production. Jessica Thomas, NO, JR, HM & CM are employed by Cochrane. GD works for Metaxis Ltd, which developed the Cochrane Register of Studies and other projects for Cochrane. James Thomas works on an online systematic review software application, EPPI‐Reviewer, which is a not‐for‐profit service of his institution. A charge needs to be made for use to contribute to its maintenance; but JT's salary is not paid for out of any income, and he makes no financial gain from the software. LB is co‐funder of the non‐profit research and innovation initiative MAGIC (Making GRADE the Irresistible Choice), which develops electronic tools and methodology for creating and publishing guidelines and evidence summaries. LB has never received any finical support from the initiative, or products coming out of it. IK is a salaried employee of Duodecim Medical Publications Ltd, a company that develops a clinical decision support service and point‐of‐care guidelines.
References
- Cochrane Informatics and Knowledge Management Department. Other software resources. tech.cochrane.org/revman/other‐resources (accessed 16 September 2014)
- Marshall C. SR Toolbox. systematicreviewtools.com (accessed 16 September 2014)
- Tsafnat G, Glasziou P, Choong MK, Dunn A, Galgani F, Coiera E. Systematic review automation technologies. Systematic Reviews 2014;3:74. www.systematicreviewsjournal.com/content/3/1/74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, Small K, Brodley CE, Lau J, Schmid CH, Bertram L, et al. Toward modernizing the systematic review pipeline in genetics: efficient updating via data mining. Genetics in Medicine 2012;14:663–669. doi.org/10.1038/gim.2012.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M TJ, O'Mara‐Eves A, Ananiadou S. Reducing systematic review workload through certainty‐based screening. Journal of Biomedical Informatics 2014; Jun 19 (published ahead of print). doi.org/10.1016/j.jbi.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cochrane Collaboration. The EMBASE screening project: six months old and going strong. www.cochrane.org/news/news‐events/current‐news/embase‐screening‐project‐six‐months‐old‐and‐going‐strong (accessed 19 September 2014)


