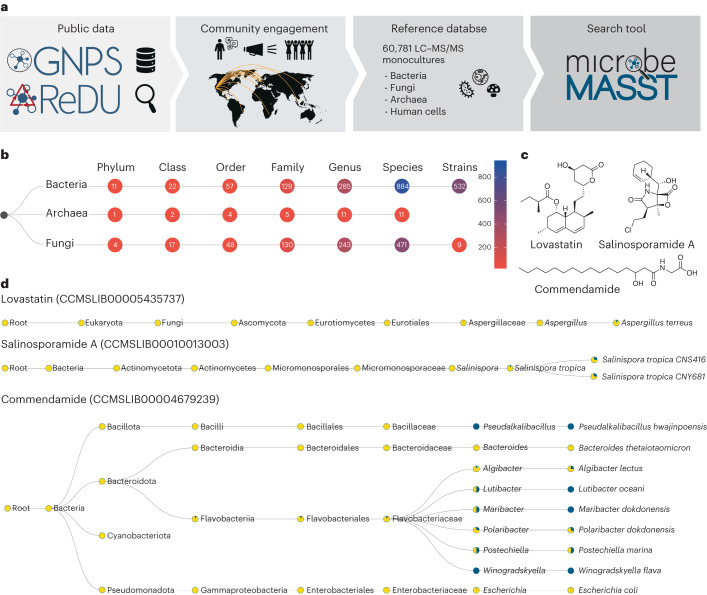
Fig. 1. The microbeMASST search tool and reference database.
a, Community contributions of data and knowledge to GNPS17, ReDU57 and MassIVE from 2014 to 2022 were used to generate the microbeMASST reference database. In addition, a public invitation to deposit data in June 2022 resulted in the further deposition of LC–MS/MS files from 25 different laboratories from 15 different countries across the globe, leading to the curation of a total of 60,781 LC–MS/MS files of microbial monoculture extracts. b, microbeMASST comprises 1,858 unique lineages across three different domains of life mapped to 541 unique strains, 1,336 species, 539 genera, 264 families, 109 orders, 41 classes and 16 phyla. c, Examples of medically relevant small molecules known to be produced by bacteria or fungi. Lovastatin, a cholesterol-lowering drug originally isolated from Aspergillus genus25; salinosporamide A, a Phase III candidate to treat glioblastoma produced by Salinispora tropica27; and commendamide, a human G-protein-coupled receptor agonist28. d, microbeMASST search outputs of the three different molecules of interest confirm that they were exclusively found in monocultures of the only known producers. Pie charts display the proportion of MS/MS matches found in the deposited reference database. Blue indicates a match with a monoculture, while yellow represents a non-match. Searches were performed using MS/MS spectra deposited in the GNPS reference library: lovastatin (CCMSLIB00005435737), salinosporamide A (CCMSLIB00010013003) and commendamide (CCMSLIB00004679239). GNPS, ReDU and microbeMASST logos reproduced under a Creative Commons license CC BY 4.0.