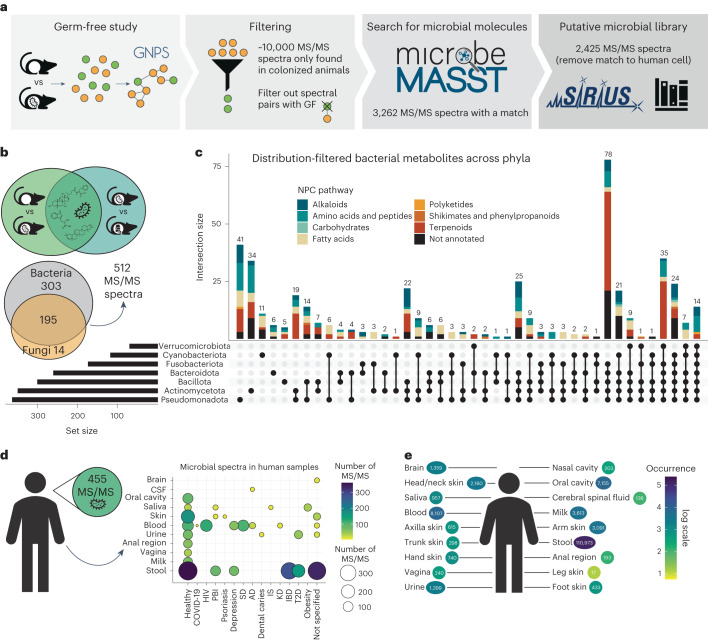
Fig. 2. microbeMASST can identify microbial MS/MS spectra within mouse and human datasets.
a, Workflow to extract microbial MS/MS spectra from biochemical profiles of 29 different tissues and biofluids of SPF mice that are not observed in GF mice30. The MS/MS spectra unique to SPF mice (10,047) were searched with microbeMASST. A total of 3,262 MS/MS spectra had a match; those MS/MS also matching human cell lines were removed, leaving a total of 2,425 putative microbial MS/MS spectra (see Methods to download .mgf file). b, The presence of the 2,425 MS/MS spectra was evaluated in an additional animal study looking at antibiotic usage40. A total of 512 MS/MS spectra, out of the 621 overlapping, were exclusively found in animals not receiving antibiotics. c, UpSet plot of the distribution of the detected MS/MS spectra (512) across bacterial phyla. Terpenoids were more commonly observed across phyla, while amino acids and peptides appeared to be more phylum specific. d, The 512 MS/MS spectra were searched in human datasets and 455 were found to have a match. These MS/MS spectra were present in both healthy individuals and individuals affected by different diseases. e, Most of the MS/MS spectra (n = 411) matched faecal samples (n = 110,973 matches), followed by blood, oral cavity, breast milk, urine and several other organs. CSF, cerebral spinal fluid; COVID-19, coronavirus disease 2019; HIV, human immunodeficiency virus; PBI, primary bacterial infectious disease; SD, sleep disorder; AD, Alzheimer’s disease; IS, ischaemic stroke; KD, Kawasaki disease; IBD, inflammatory bowel disease; T2D, type II diabetes. GNPS and and microbeMASST logos reproduced under a Creative Commons license CC BY 4.0; SIRIUS logo reproduced under a Creative Commons license CC BY 4.0-ND.