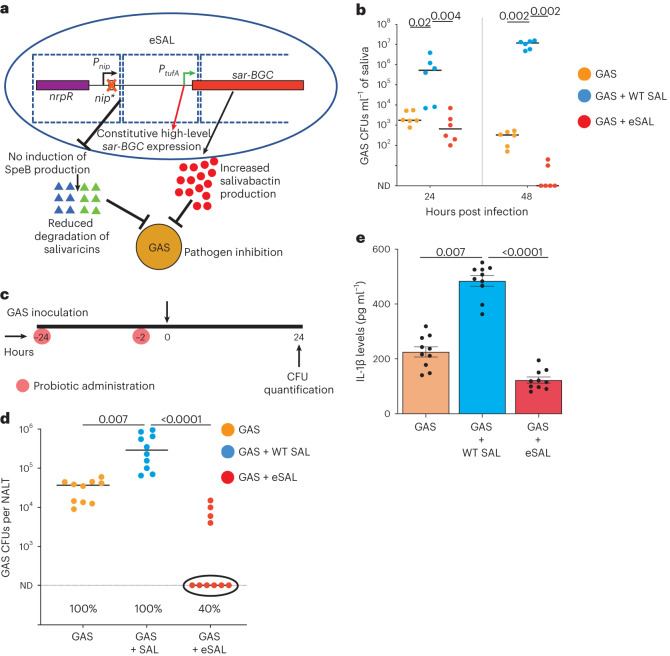
Fig. 6. Inhibition of GAS colonization by an engineered probiotic.
a, Schematics of genetic modifications in engineered S. salivarius (eSAL). The nip* mutation was introduced to abolish NIP production in S. salivarius and early induction of SpeB production in GAS during dual species growth. Transcription of sar operon was coupled with constitutively active PtufA promoter that drives high-level sar BGC expression. Delayed SpeB protease production is likely to disarm pathogen defence and result in increased salivaricin levels due to reduced degradation by SpeB. Collectively, increased salivaricin and salivabactin levels may lead to improved pathogen inhibition and clearance of GAS in the host. b, GAS clearance from saliva by engineered probiotic (eSAL) as assessed by CFU analyses. c, Experimental design to assess the probiotic efficacy of eSAL in vivo. Each group (n = 10 mice per group) received 108 CFUs of either WT SAL or eSAL intranasally at the indicated timepoints. One day after the first dose of probiotic administration, single dose (108 CFUs) of GAS was given intranasally. GAS burden was assessed by CFU analyses. d, eSAL was more efficacious than WT SAL in preventing GAS colonization in mouse nasopharynx. The circles indicate the lack of detectable GAS colonies in eSAL-treated group. The numbers below indicate the percentage of animals colonized by GAS in each group. e, IL-1β levels in nasopharyngeal swabs as assessed by ELISA. In b and d, ND indicates limit of detection. In b, the detection limit was set at zero as GAS was cleared from saliva in eSAL-treated group, whereas in d, ND was set at 100. In b and d, data represent the geometric mean from three independent experiments that was analysed in duplicate. In e, data are presented as mean ± s.e.m. Statistical significance was analysed by Kruskal–Wallis test. In b, d and e, GAS + WT SAL group was used as a reference.